Méthodes pour l’étude de structures, dynamiques et interactions d’acides nucléiques non canoniques
2025-12-19
Career Overview
Professional Journey
- M2 Intern - LSOSCO - 2008
- UMR 8123 CNRS-UCP-ESCOM
- PhD Student - U. Paris XI - 2008-2012
- UMR176 CNRS, Institut Curie
- Postdoc - UBC - 2012-2013
- Chemistry Dpt
- Postdoc - IECB - 2013-2015
- U869 CNRS/U. Bordeaux
- Research Scientist - Quality Assistance - 2015-2017
- R&D Dpt
- MCU - University of Bordeaux - 2017-present
- ARNA - INSERM U1212, CNRS UMR 5320, U. Bordeaux
Teaching & support duties
Topics
- Analytical Chemistry
- Regulatory aspects
- Biomolecules
- Scientific communication
- Data processing and visualization
Students
- Health Sciences
- Pharmacy (L2/L3)
- DEUST (Production, contrôle et qualité des produits de santé, L1/L2)
- TECSAN (Technologies pour la santé,L3)
- Master IPPS (Industries Pharmaceutiques et Produits de Santé, M1)
- Master DACQ (Développement Analytique et Contrôle Qualité, M2)
- CHU Bodeaux
- Préparateurs en pharmacie hospitalière
- Collège des écoles doctorales
- PhD students (all fields)
- Reference person @ ARNA for
- Research data; member of the Bordeaux Data Workshop
- Open Science
- Manuscript: hdr.largy.fr
- Slides: slides.hdr.largy.fr (source code)

PhD at the Institut Curie/CNRS/University Paris XI

A quick introduction on G-quadruplexes

A quick introduction on G-quadruplexes

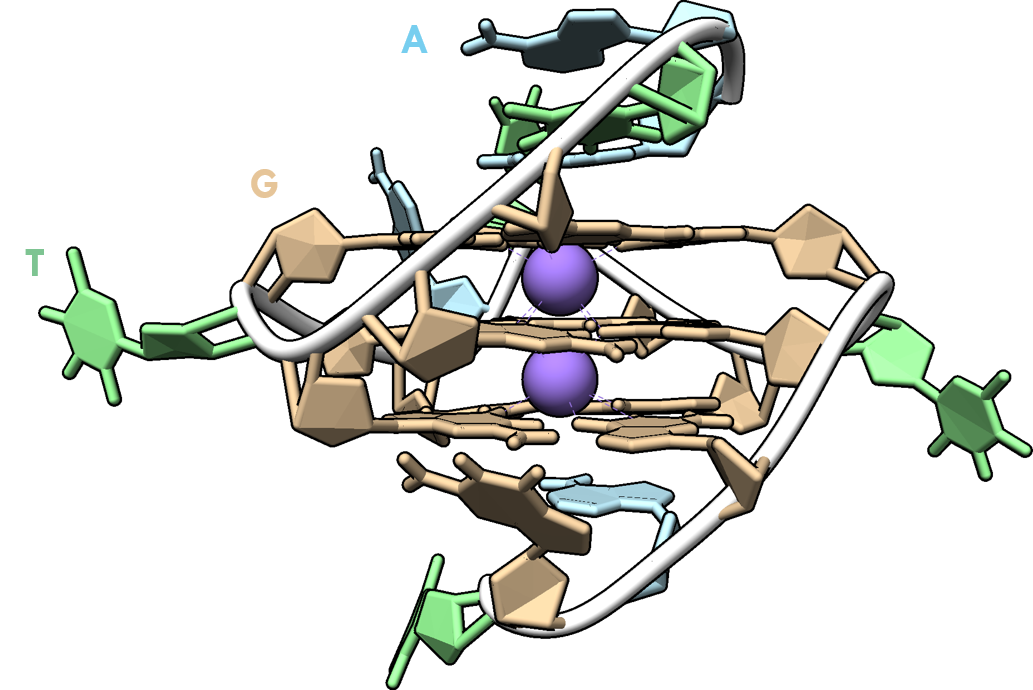
Moving targets
- Drug targets…
- Involved in biological processes at DNA/RNA levels
- Structurally different from duplexes
- …and drugs?
- Some aptamers are or contain a G4
- Very polymorphic
- Can fold into different conformers in equilibrium
- May not be fully folded
- Very dependent on cation concentration/nature
- Goal: selective binding to modulate biological functions
- Generally, \(\pi\)-stacking ligands on external tetrads
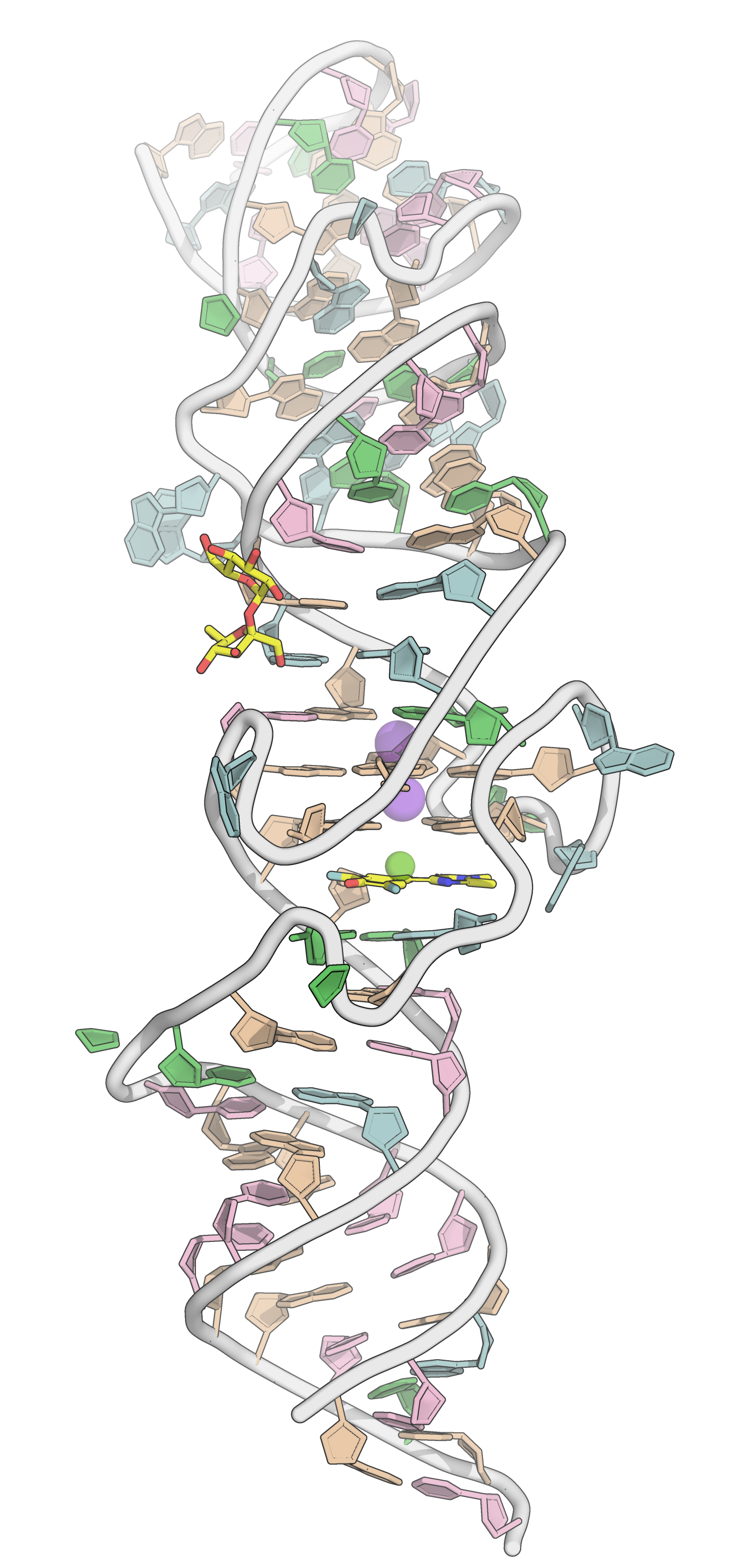

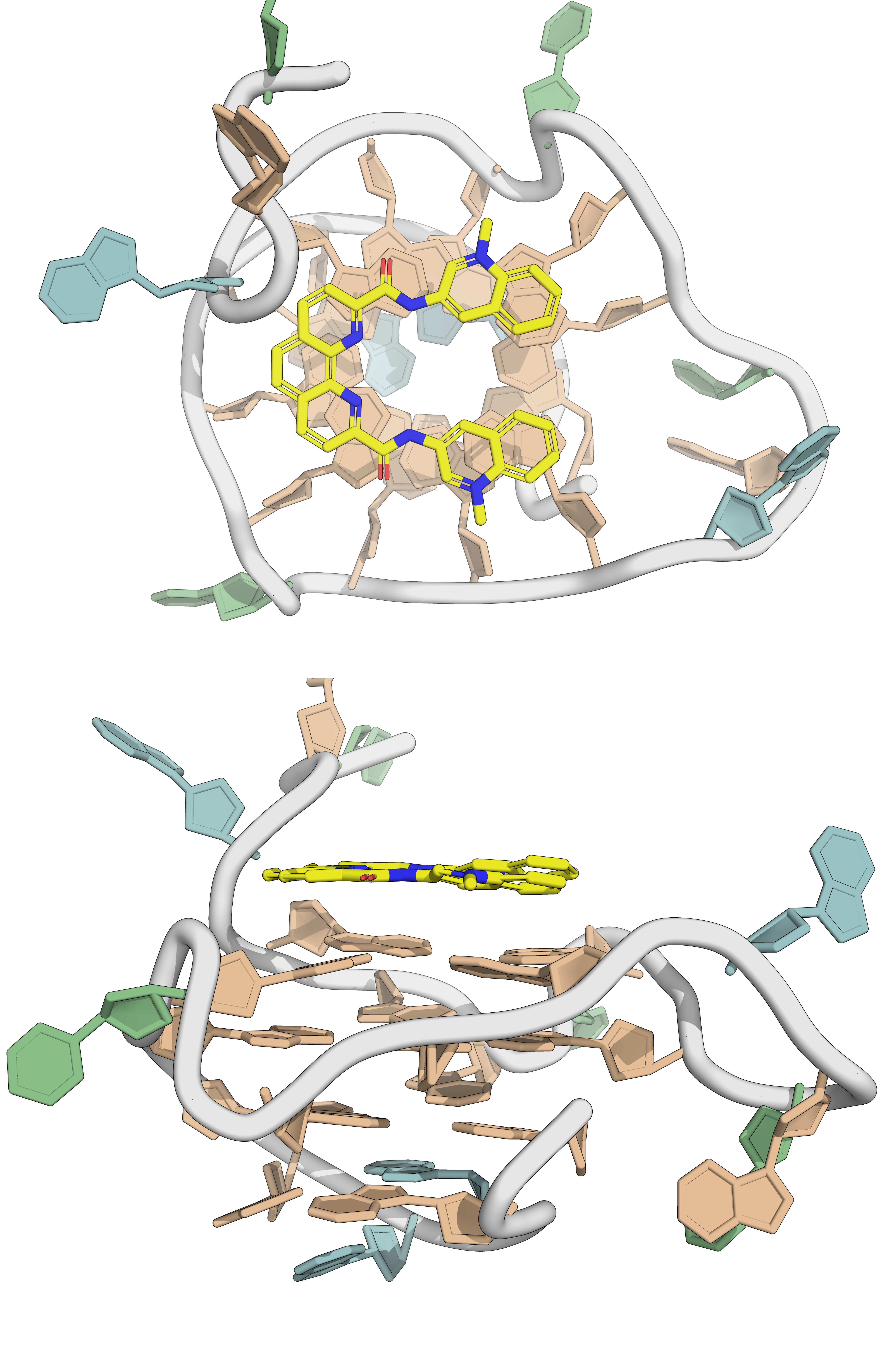
PDB: 4TS2: Warner et al., Nat. Struc. Mol. Biol., 2014, 21, 658
Largy et al., Chem. Rev. 2022, 122, 8, 7720
Largy, E., Mergny, J.-L. et Gabelica, V. Role of Alkali Metal Ions in G-Quadruplex Nucleic Acid Structure and Stability. 2016, p.203‑258.
Goals of the PhD project

- Initially
- Synthesize metal complexes as G4 ligands
- Understand the influence of metal ions and aromatic ligand on binding selectivity.
- Synthesize ligands with an added value
- Topological selectivity
- Original binding mode
- Functionalization: fluorescence, biotinylation, halogenation,…
- Develop analytical tools to
- Characterize G4/ligand interactions
- Discover new G4 ligands
Axis 2. Methods for G4·ligand interaction characterization
Fluorescent Interacalator Displacement Assay (FID)
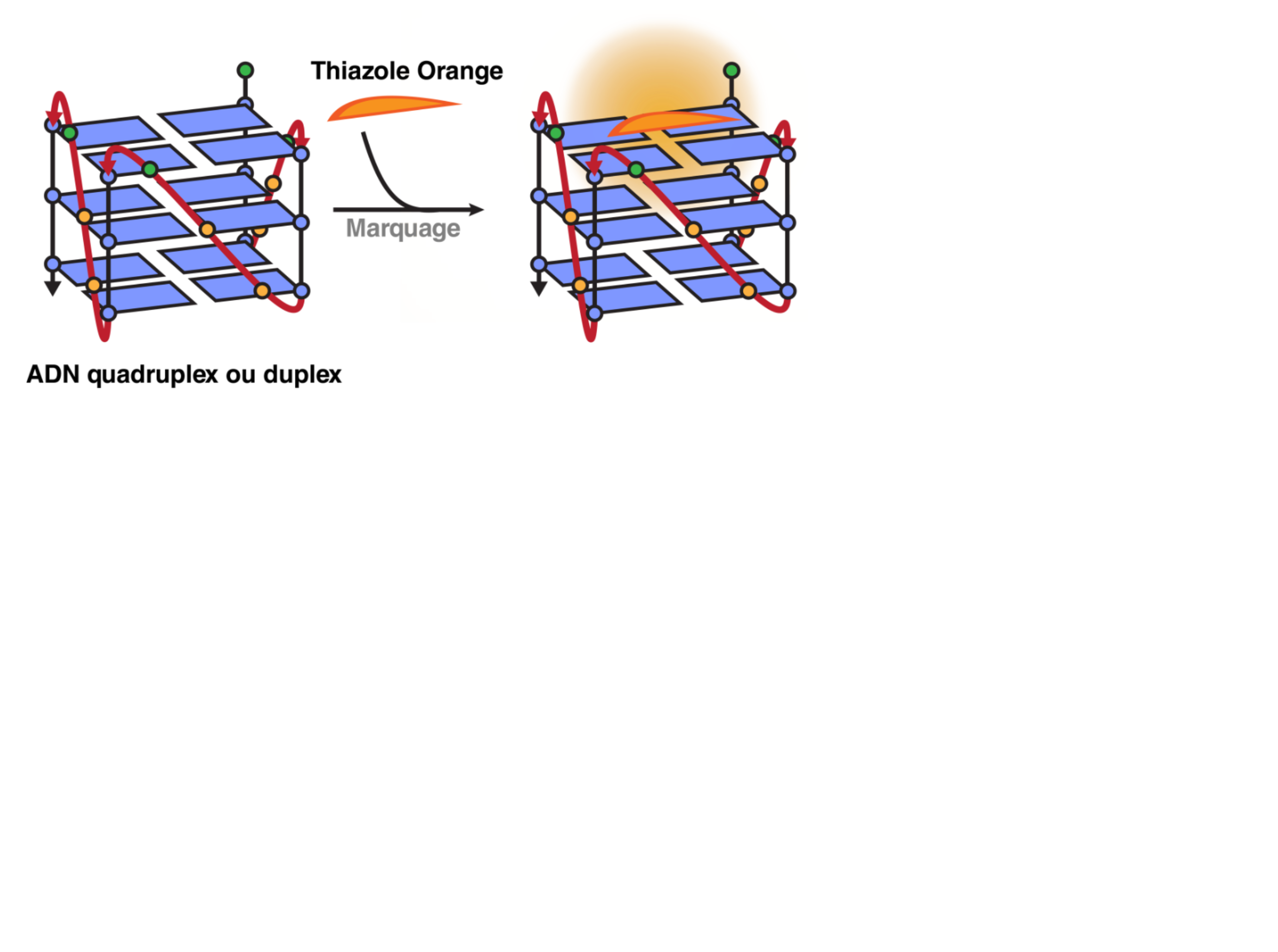
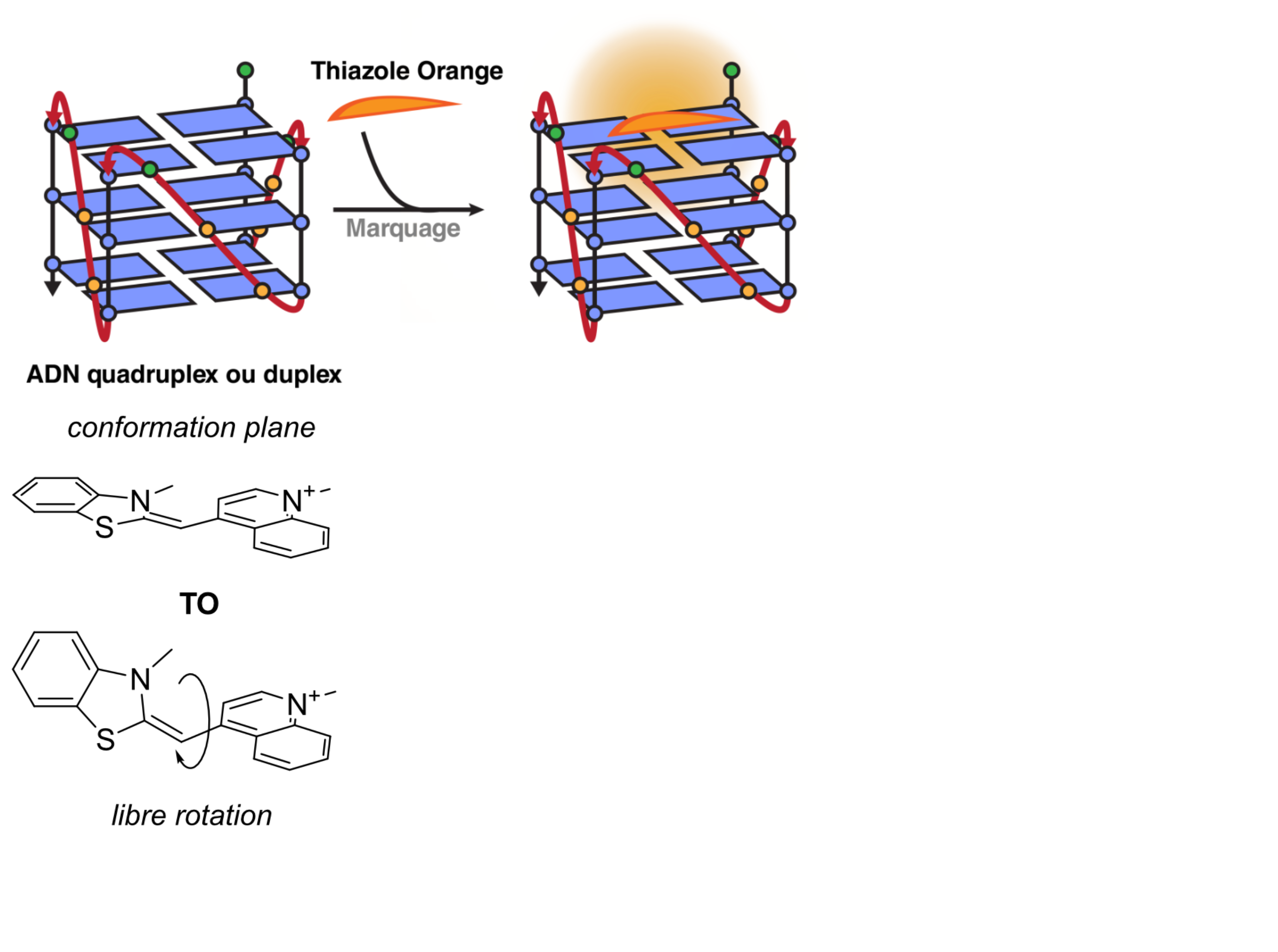
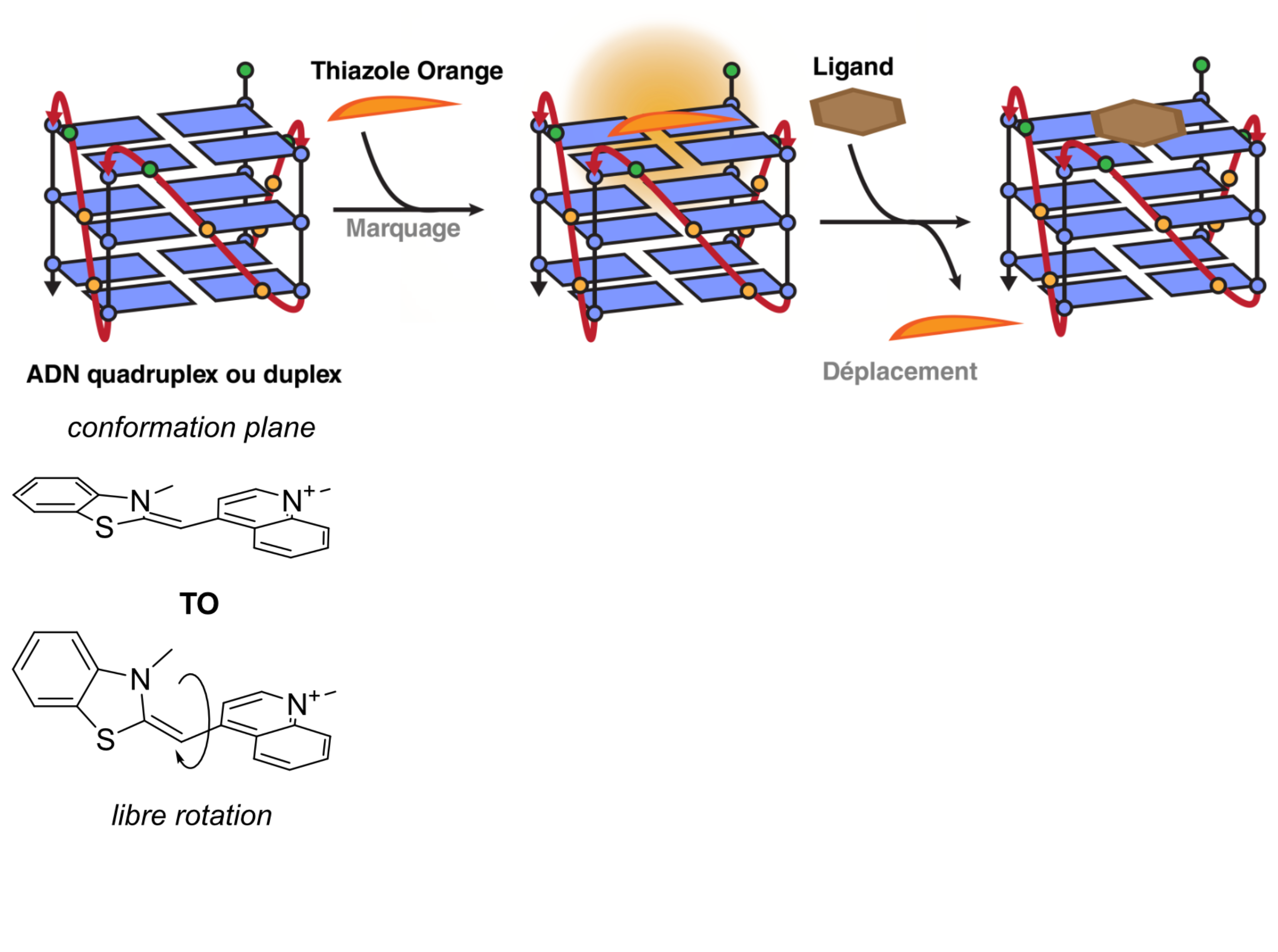
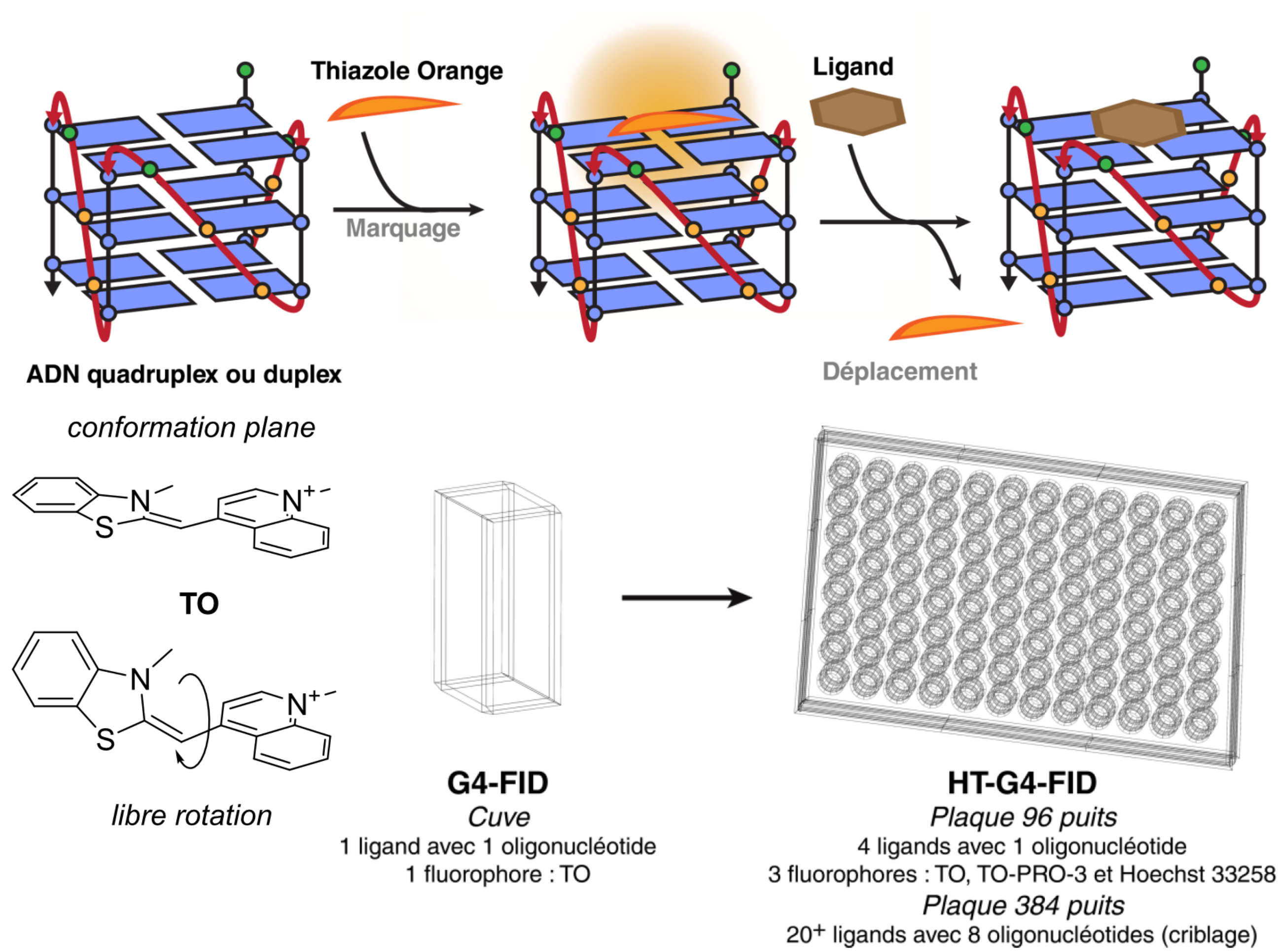
Largy, E., Hamon, F. et Teulade-Fichou, M.-P. Anal. Bioanal. Chem., 2011, 400, 3419.
Tran, P.L.T., Largy, E., Hamon, F., Teulade-Fichou, M.-P. et Mergny, J.-L. Biochimie, 2011, 93, 1288
FID examples: DNA mismatch recognition
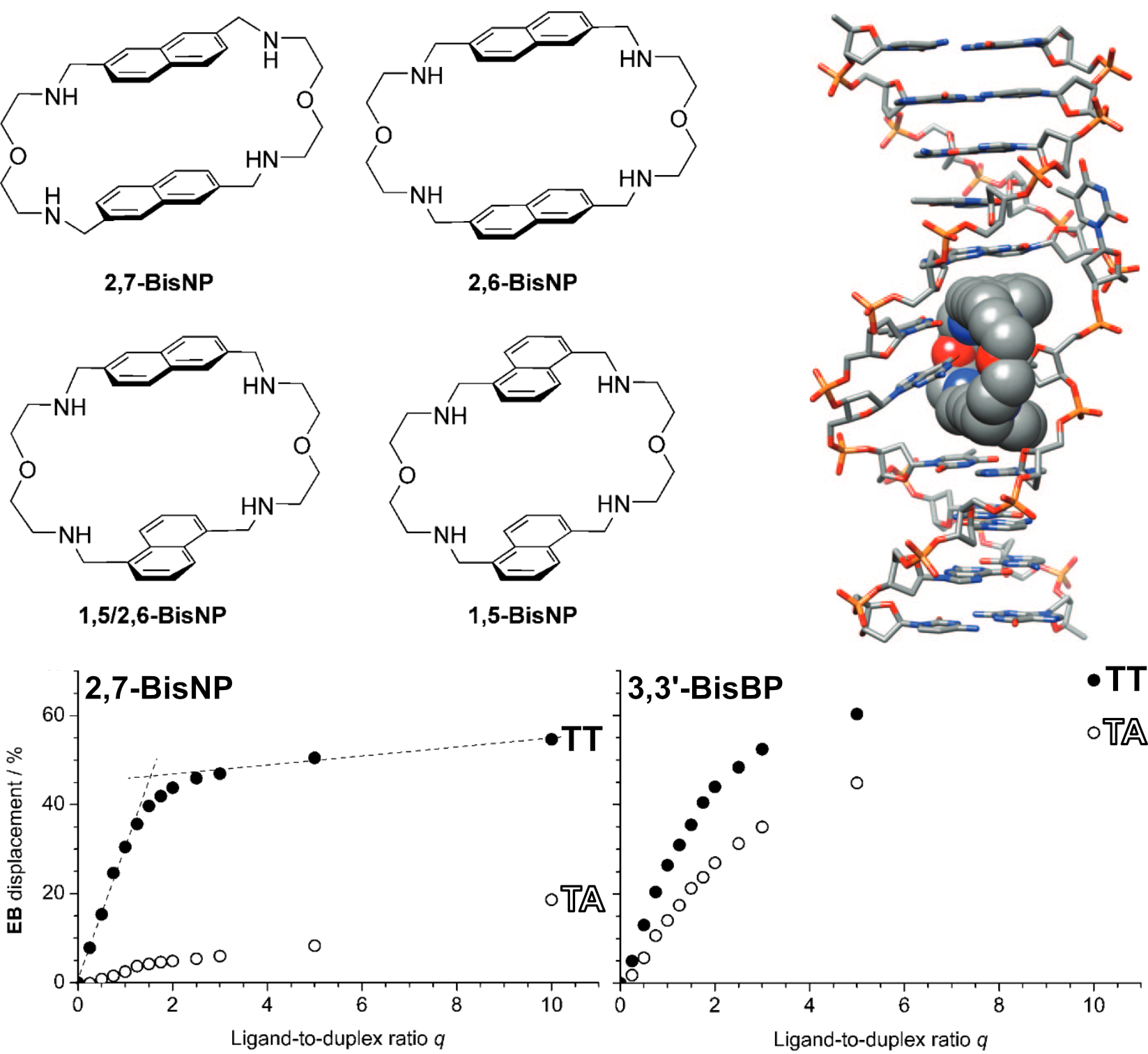

Granzhan, A., Largy, E., Saettel, N. et Teulade-Fichou, M.-P. Chem. Eur. J., 2010, 16, 878.
FID examples: RNA binding affinity
Halder, K., Largy, E., Benzler, M., Teulade-Fichou, M.-P. et Hartig, J.S. ChemBioChem, 2011, 12, 1663
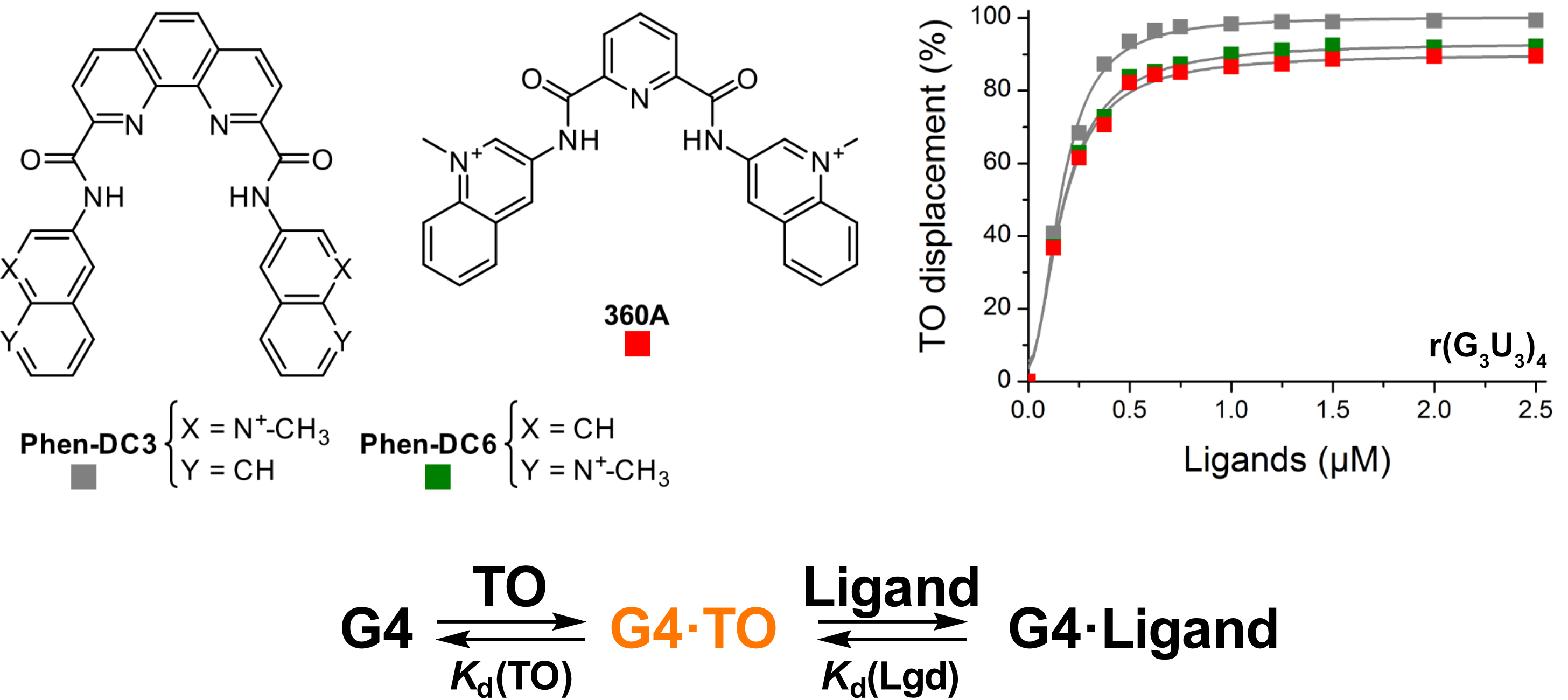
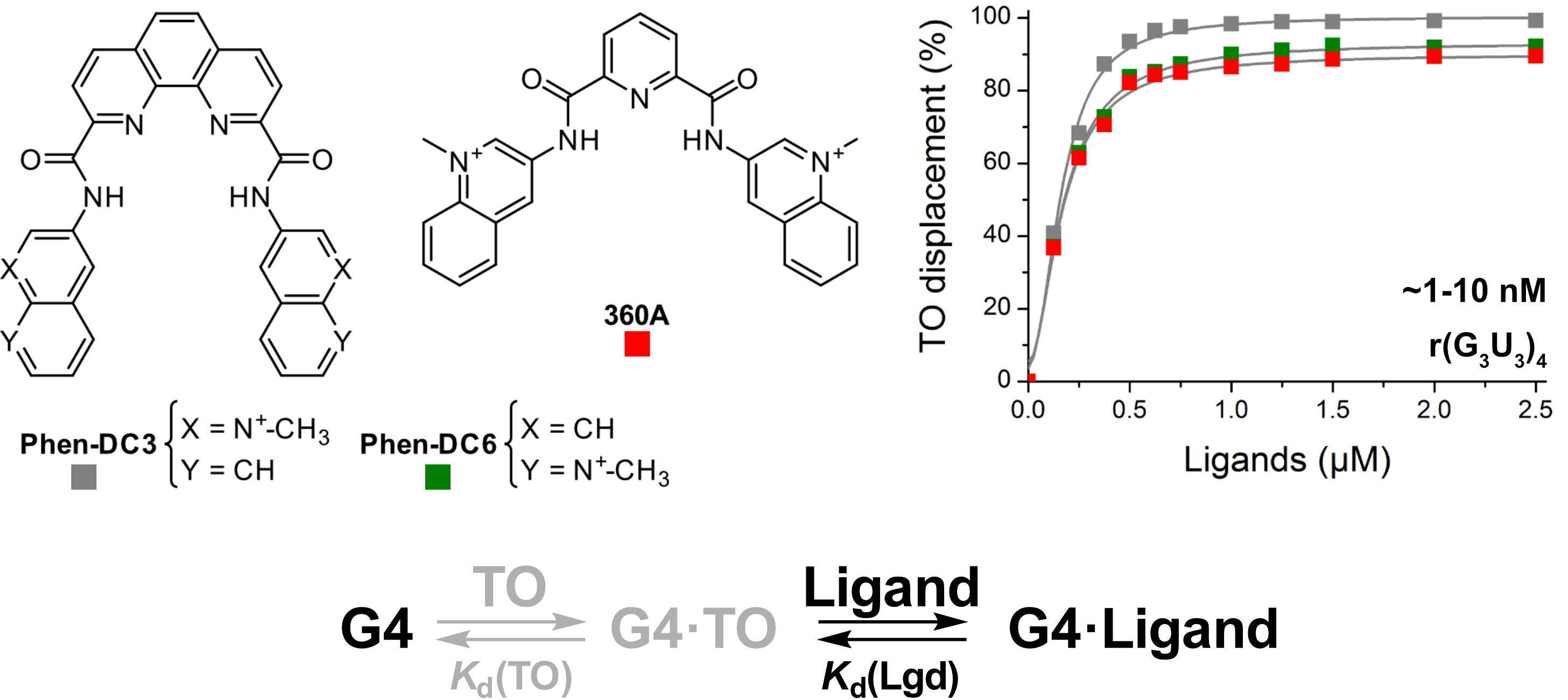
Screening of G4 ligands from a chemical library
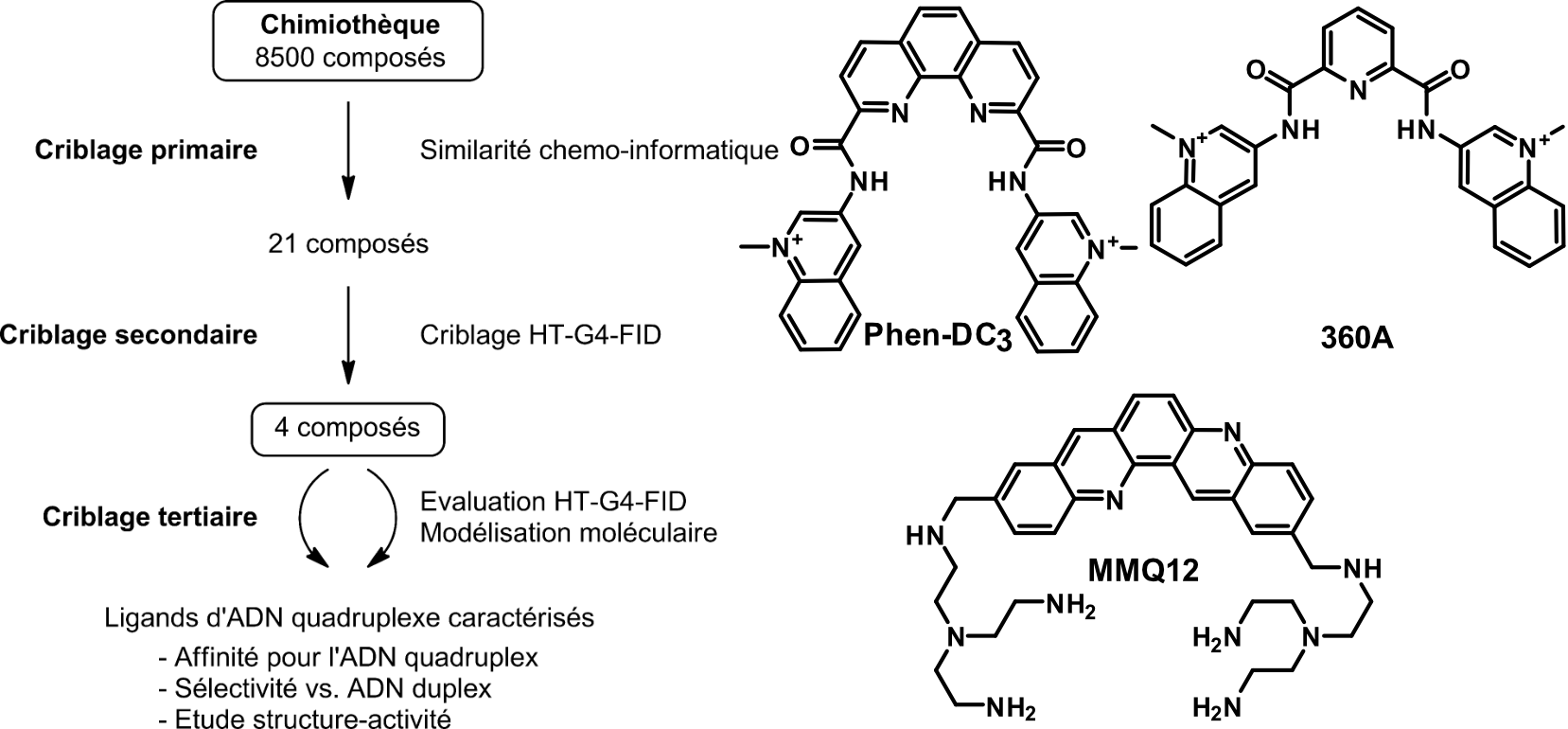
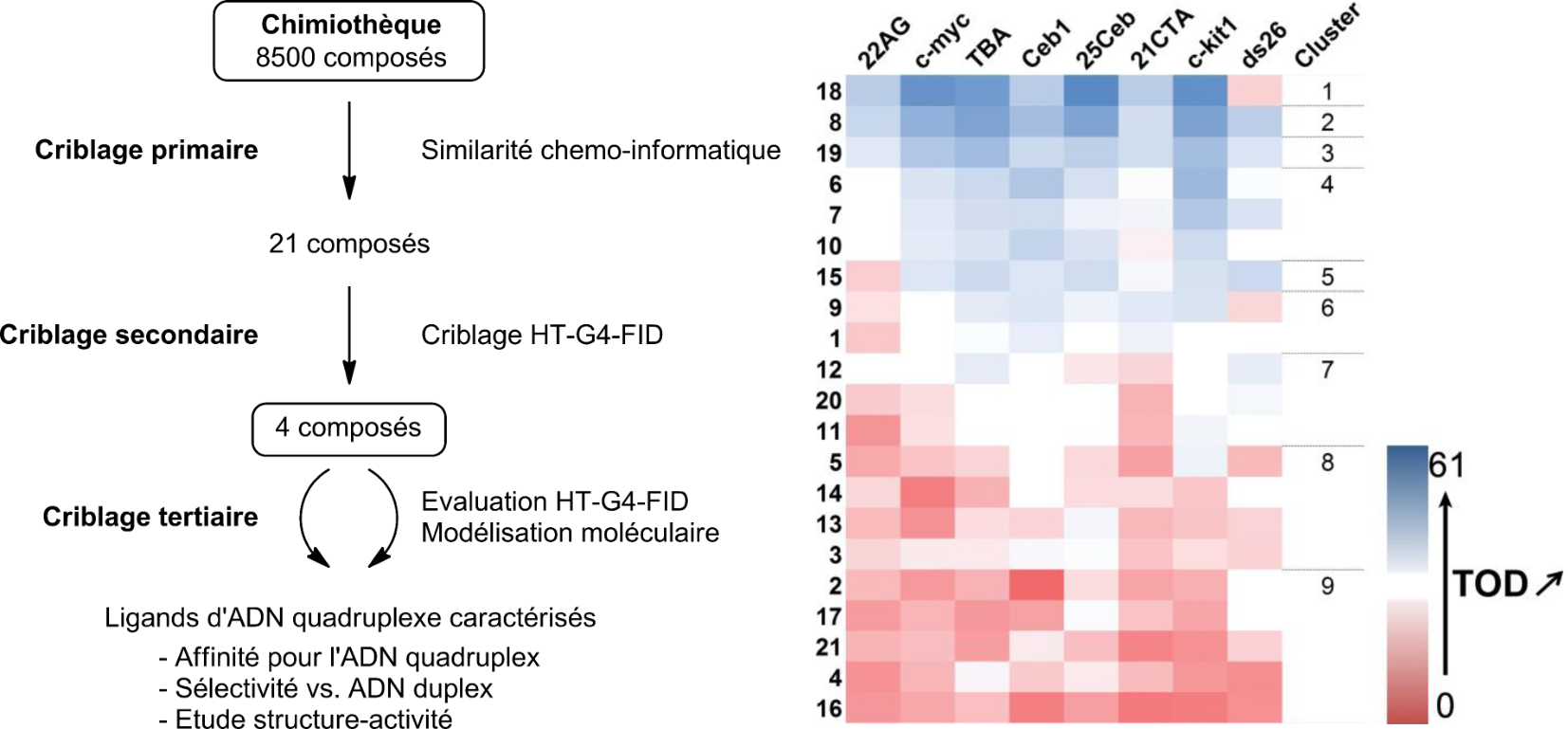
Largy, E., Saettel, N., Hamon, F., Dubruille, S. et Teulade-Fichou, M.-P. Curr. Pharm. Des., 2012, 18, 1992
Largy, E. et Teulade-Fichou, M.-P. Screening for Quadruplex Binding Ligands: A Game of Chance? 2012
Axis 1. Synthesis of G4 ligands with added value
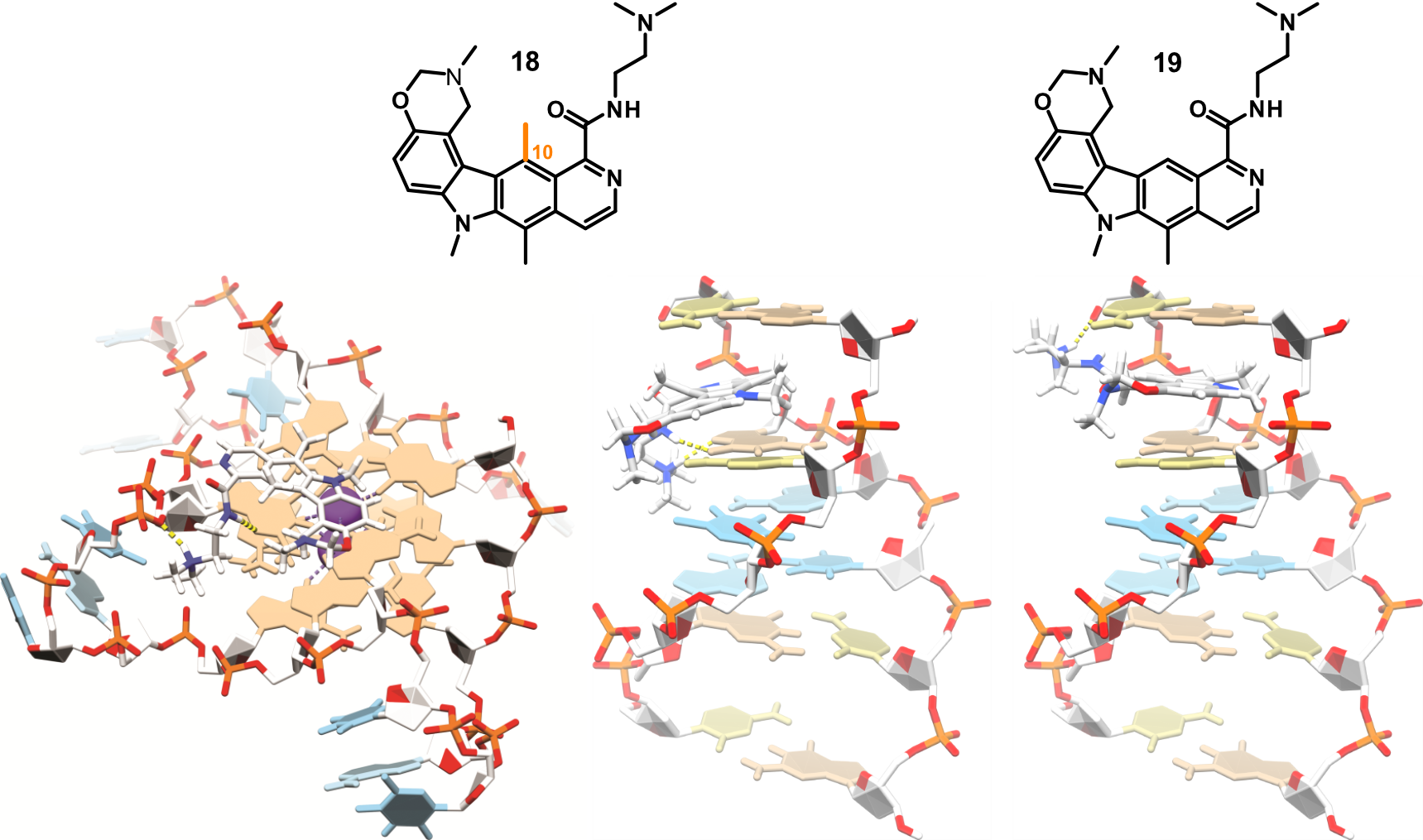
Metal complexes as covalent G4 binders

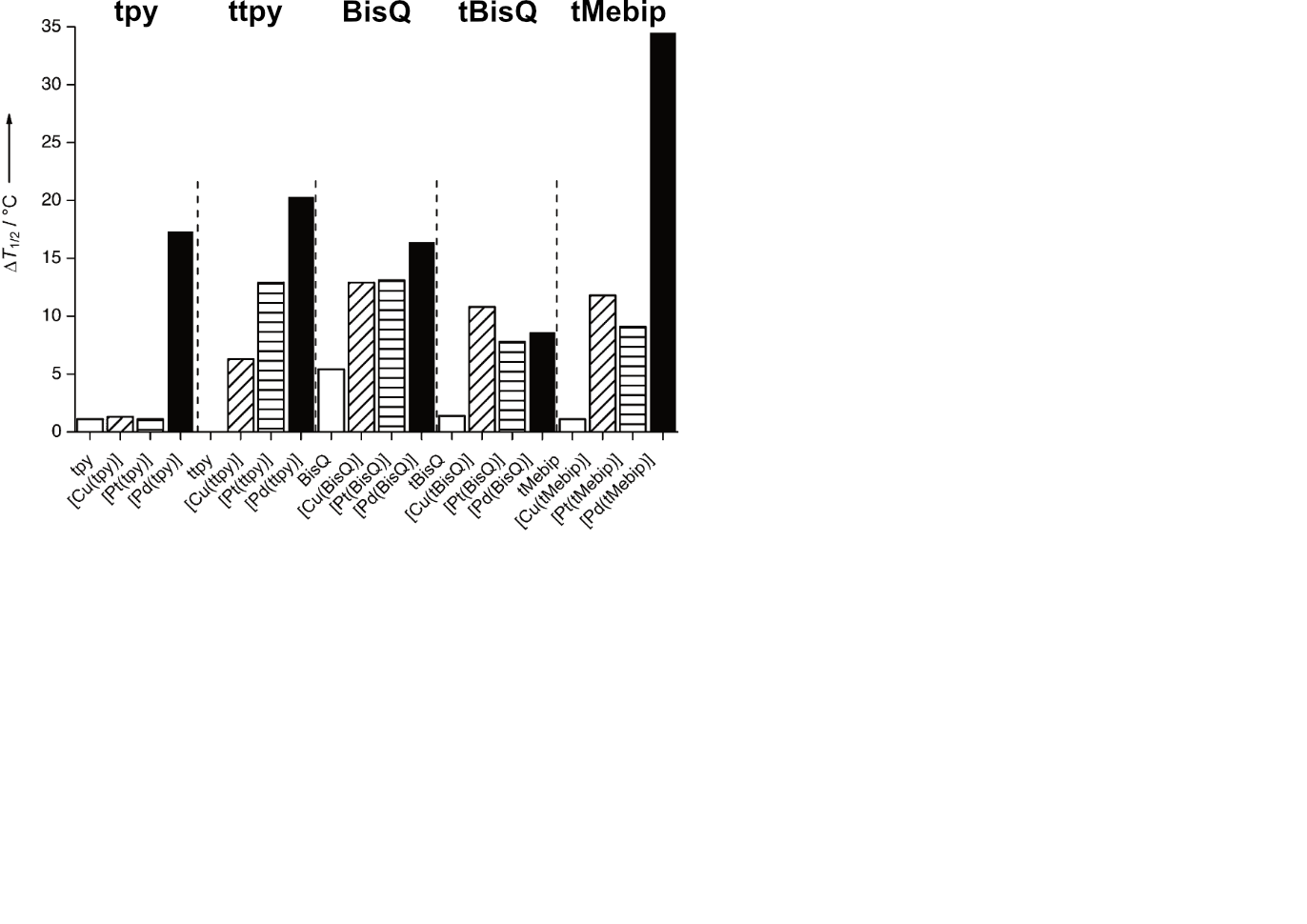
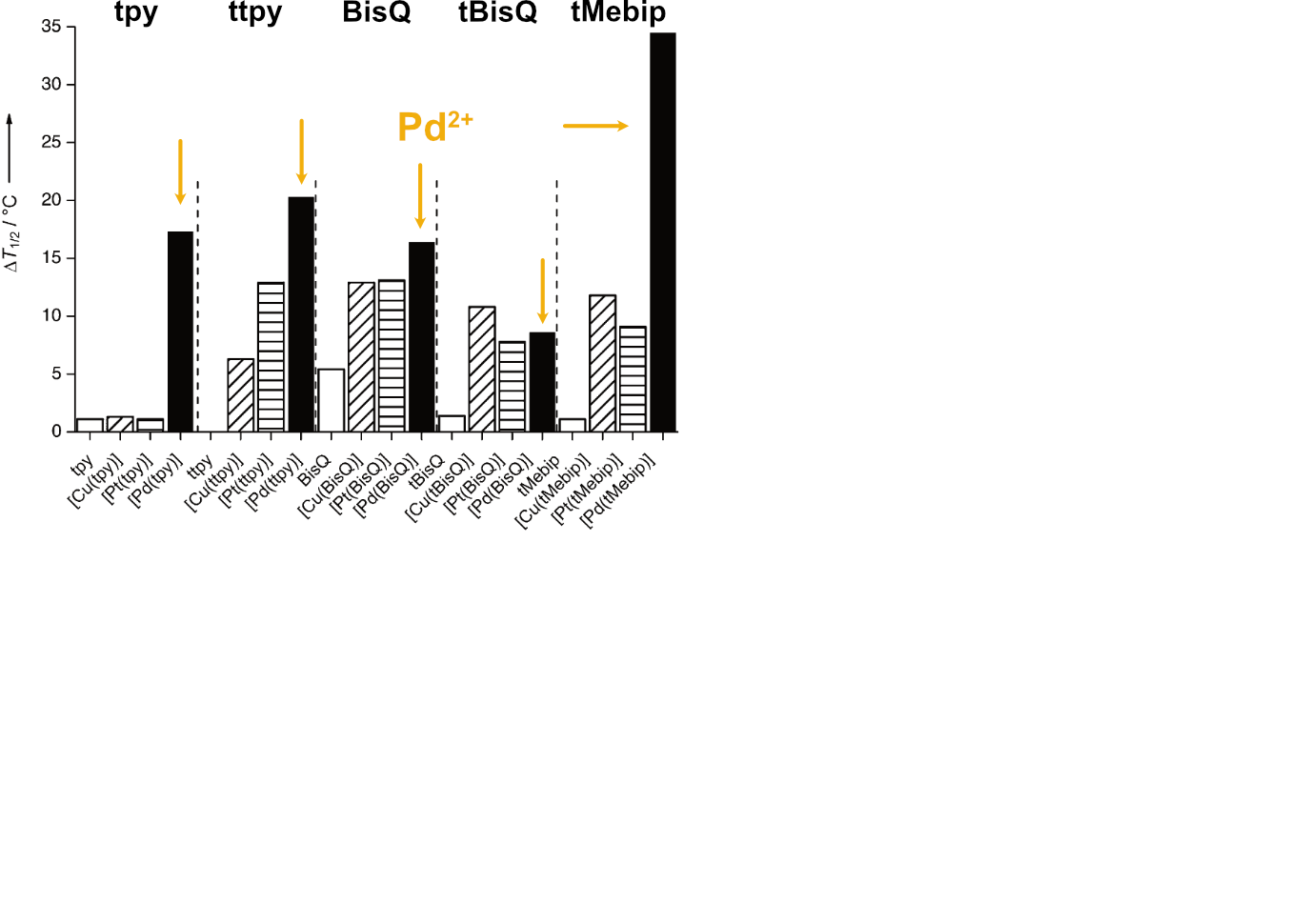
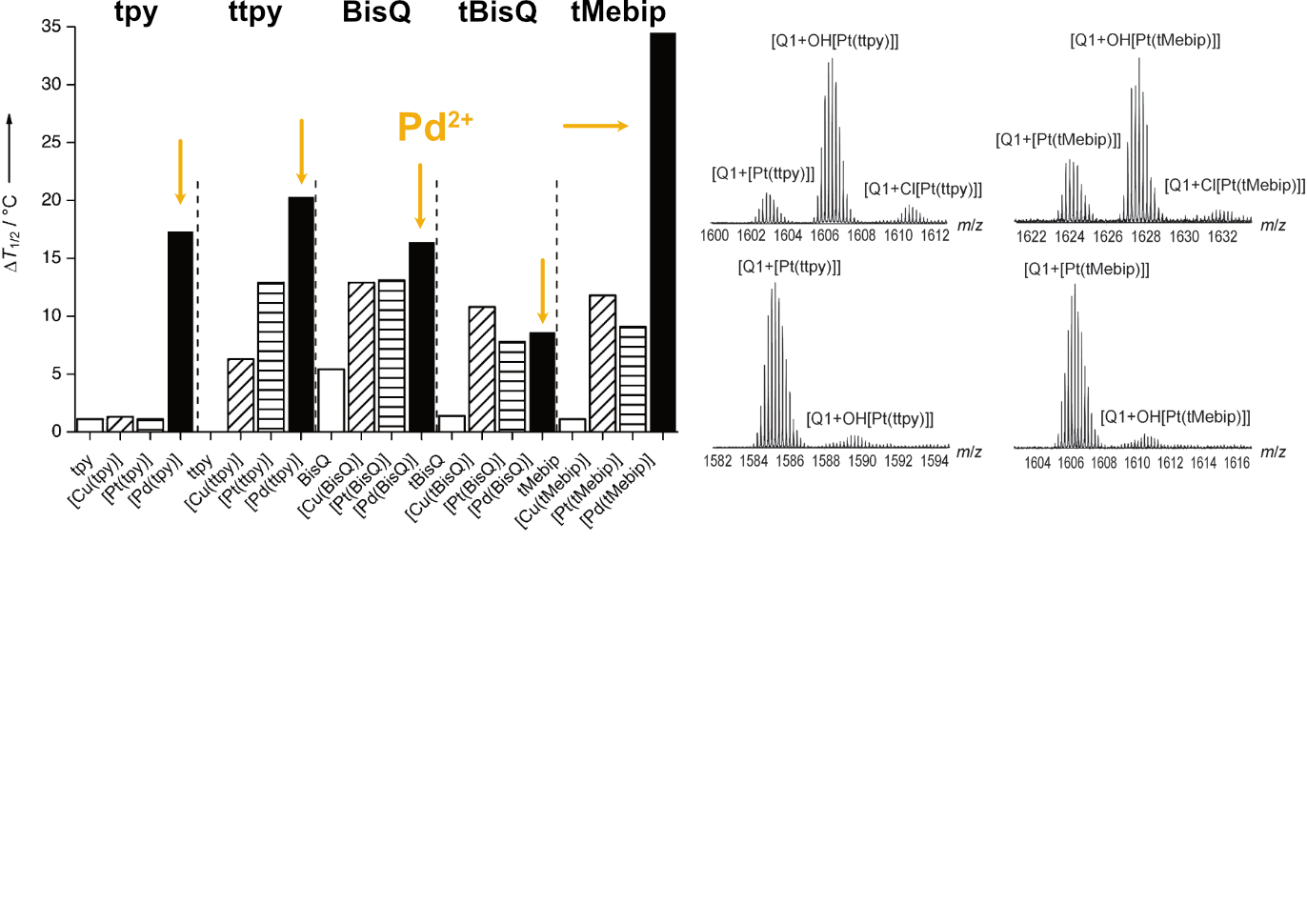
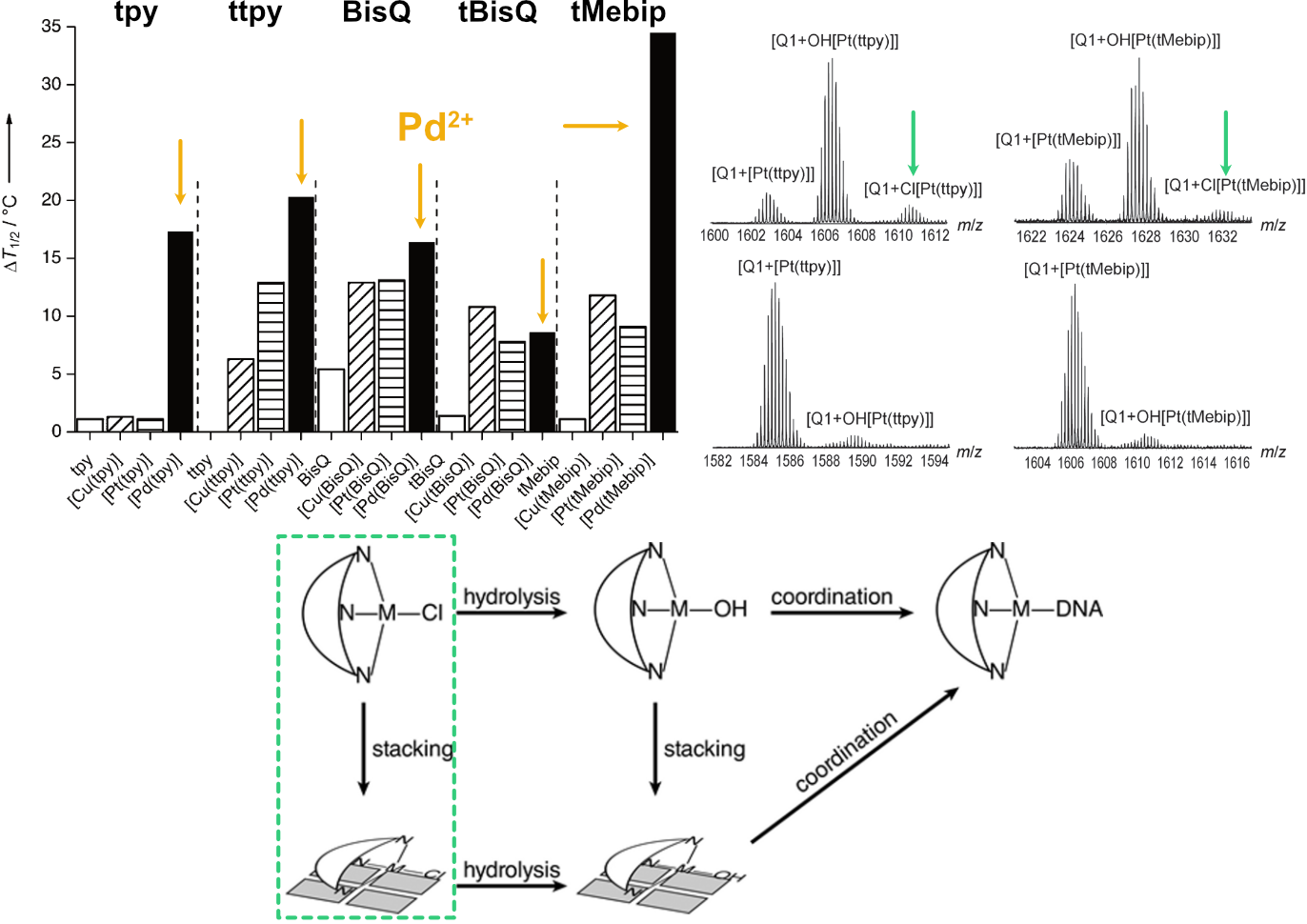
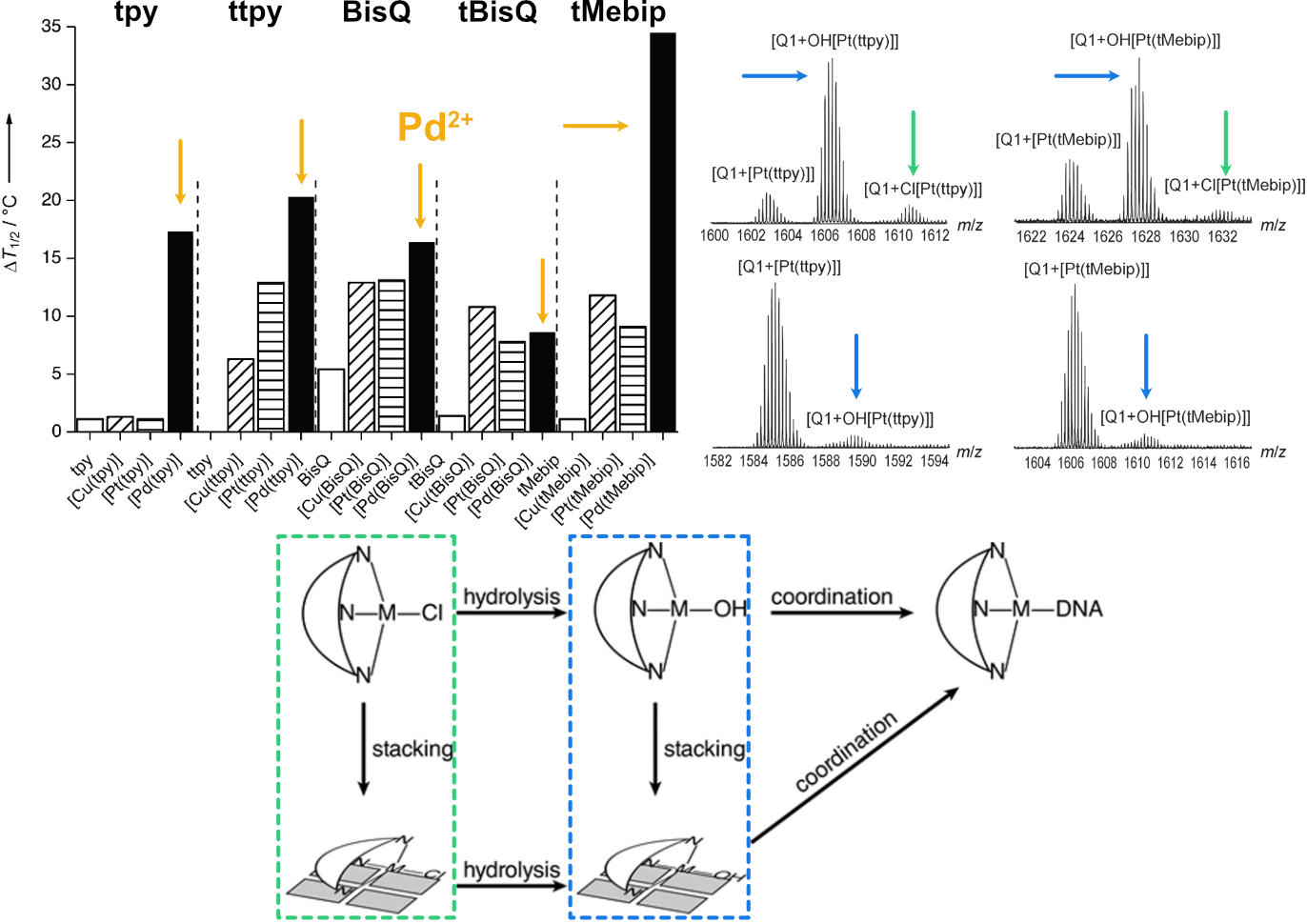
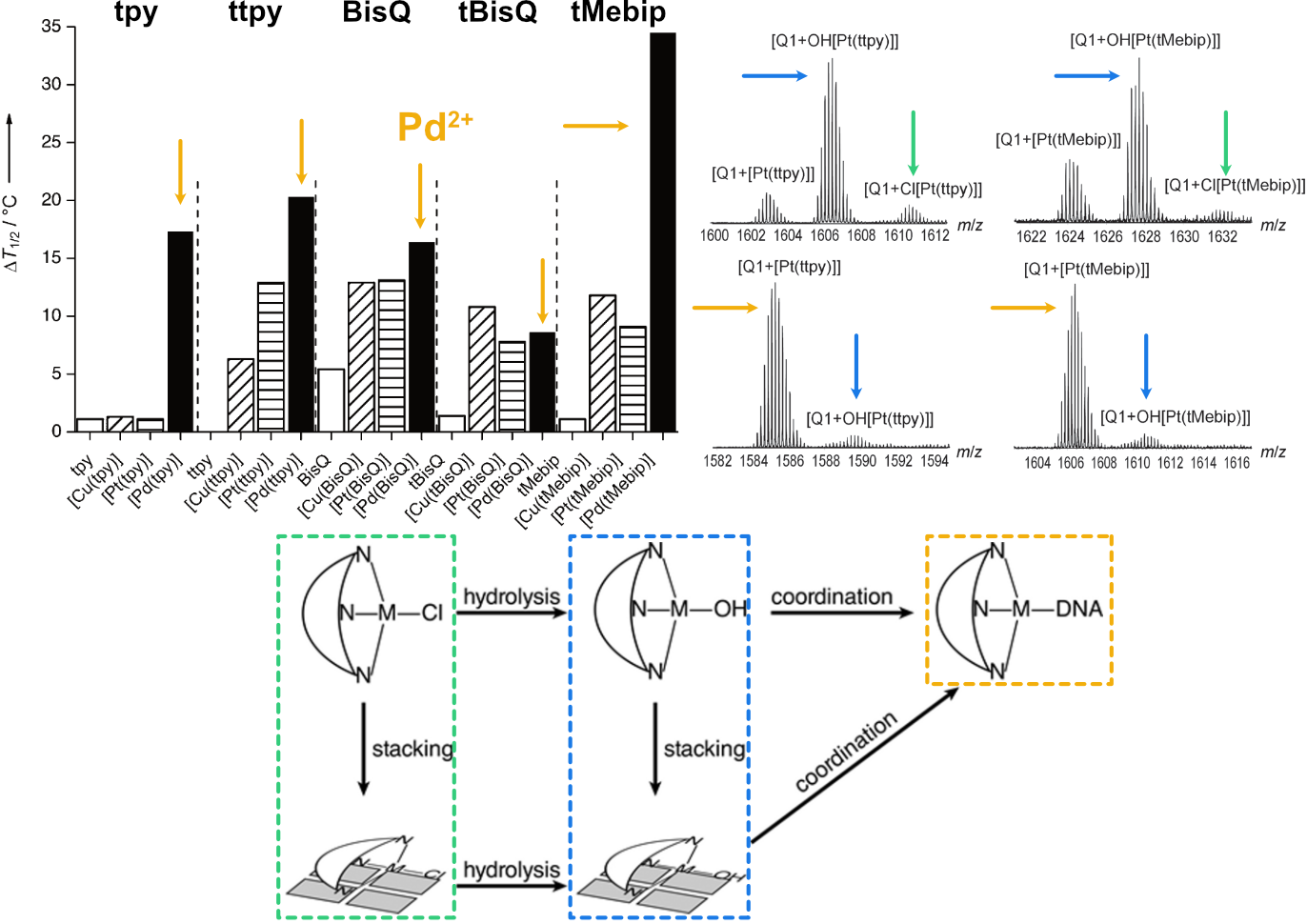




Largy, E., Hamon, F., Rosu, F., Gabelica, V., DePauw, E., Guédin, A., Mergny, J.-L. et Teulade-Fichou, M.-P. Chem. Eur. J., 2011, 17, 13274
Acyclic oligoheteroaryl ligands as groove binders(?)
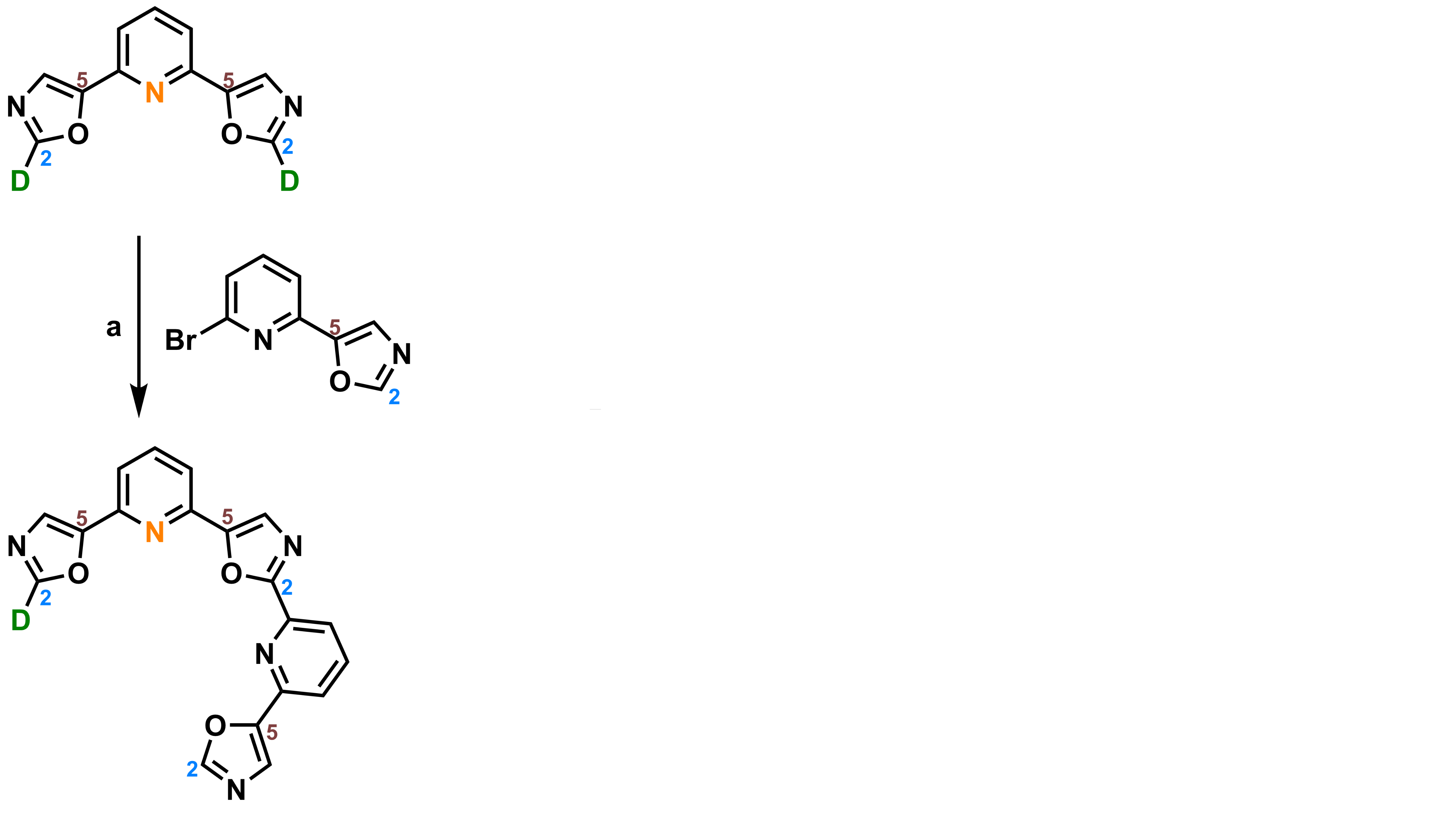
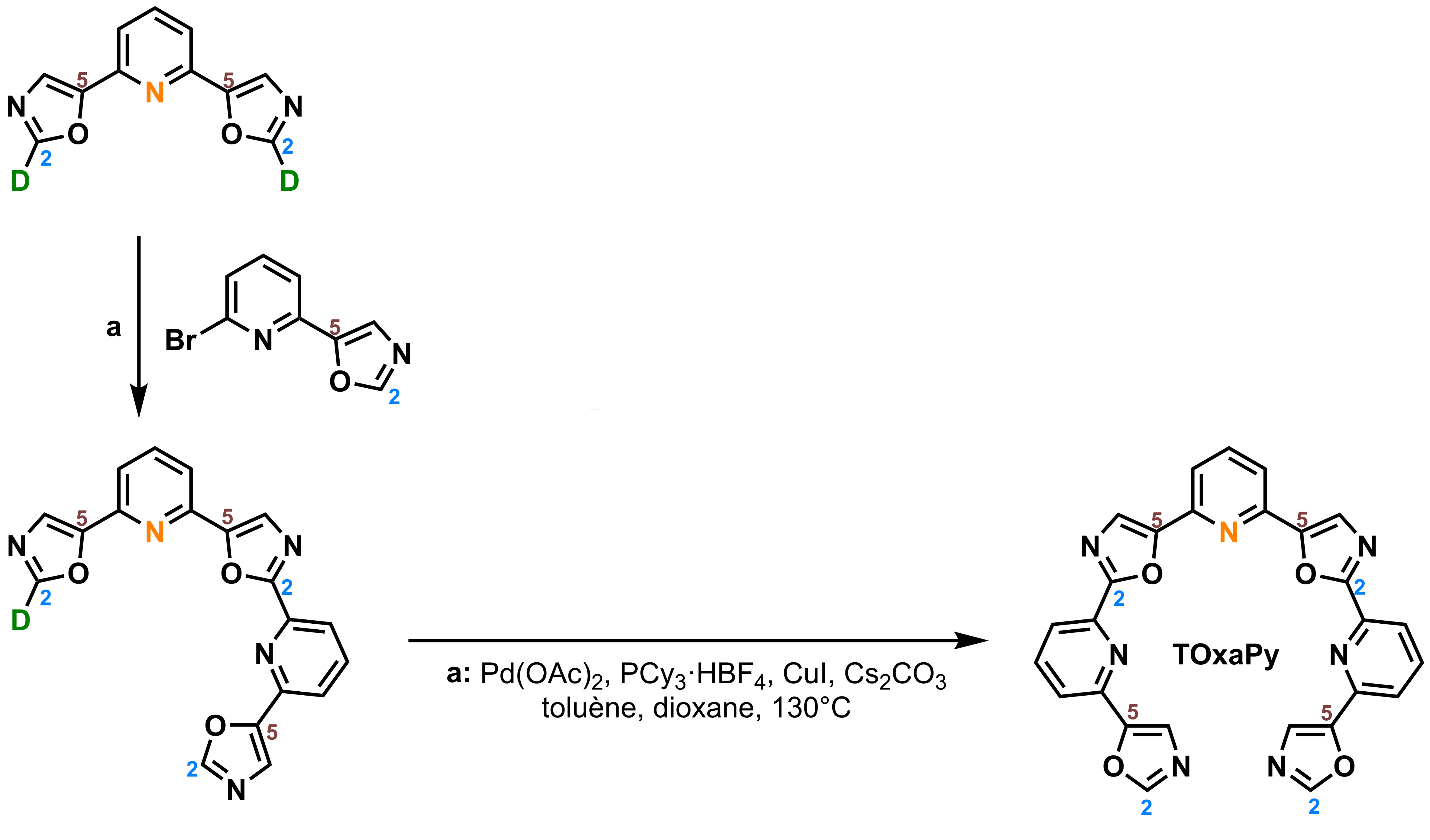
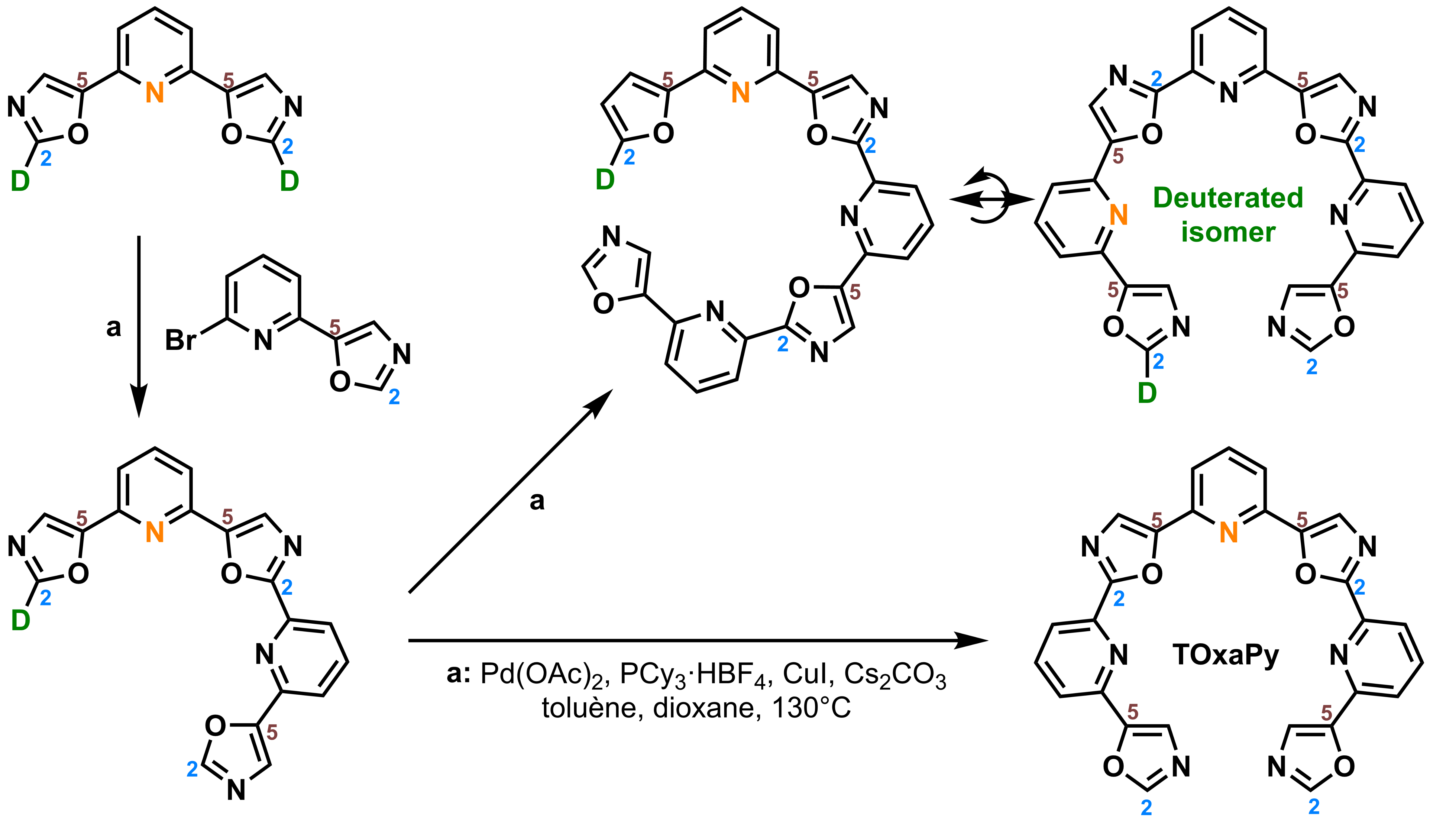
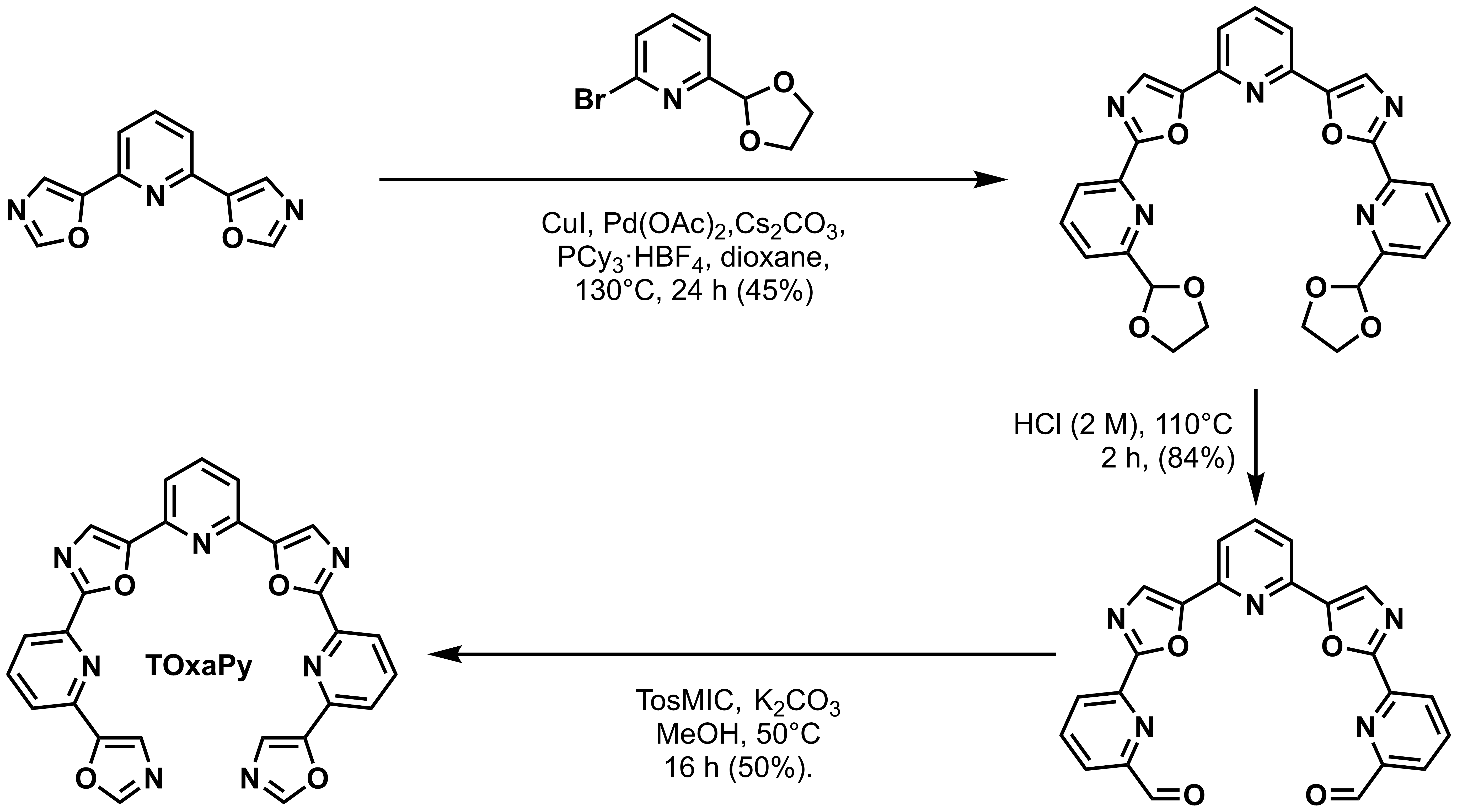

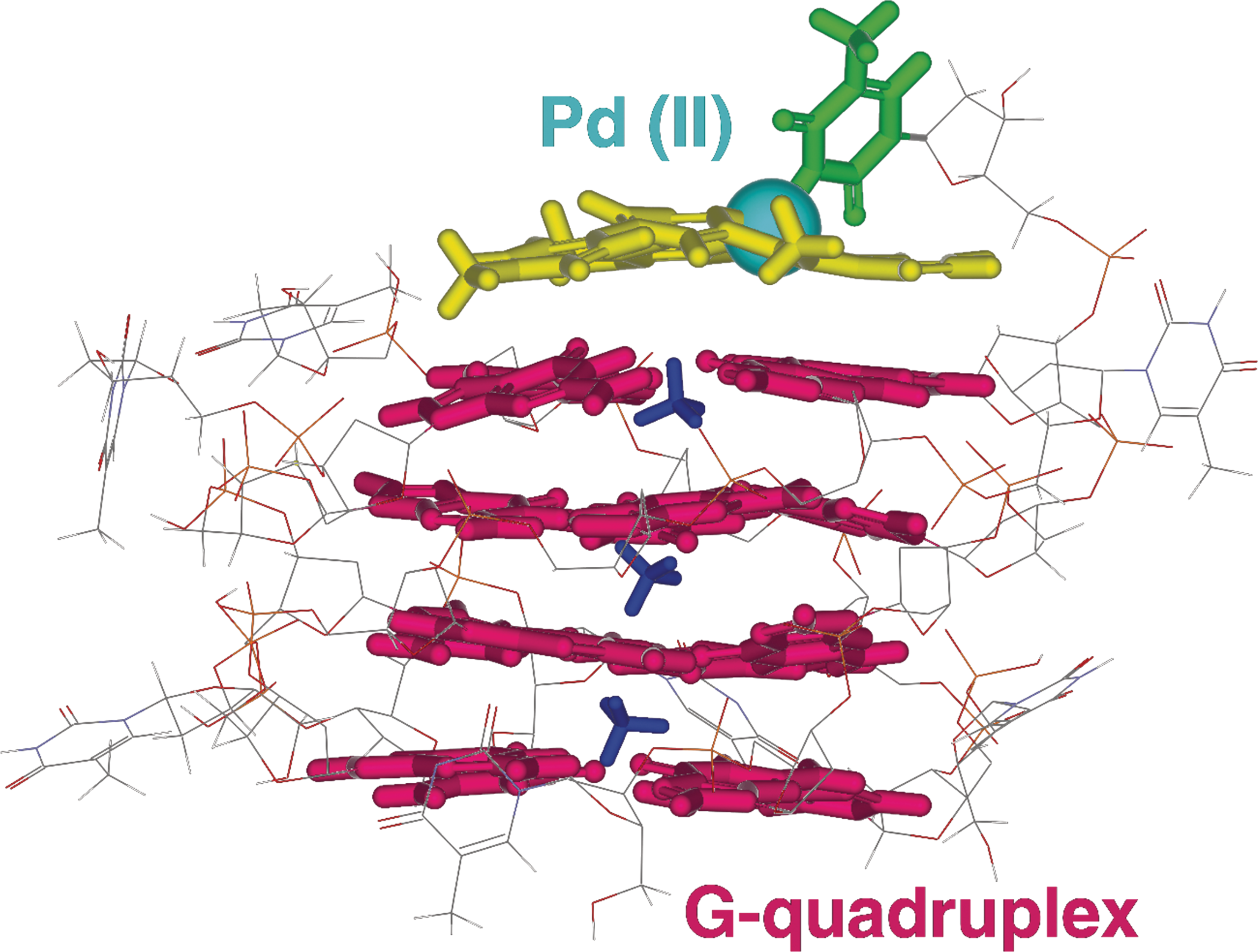
Hamon, F., Largy, E., Guédin-Beaurepaire, A. […], Teulade-Fichou, M.-P. Angew. Chem. Int. Ed., 2011, 50, 8745
Acyclic oligoheteroaryl ligands as groove binders(?)





Hamon, F., Largy, E., Guédin-Beaurepaire, A. […], Teulade-Fichou, M.-P. Angew. Chem. Int. Ed., 2011, 50, 8745
Postdoc at the University of British Columbia

Invading duplexes with bifacial nucleosides
- Antisense strategy limited
- Requires strand displacement
- Not particularly energetically favorable
- Energetic gain by forming additional H-bonds
- Exploit H-bond donor/acceptor groups on both strands
- First generation JAT
- Donor/Acceptor network designed to target bp
- Suboptimal protonation state at physiological pH
- \(pK_{a} \approx 7.5\)
- Second generation NJAT
- Lowered pKa
- Improved binding
- Sequence-selective (Tm ~ 10°C > G/C)
- Adequate base-pairing geometry
- No significant distortion of duplex
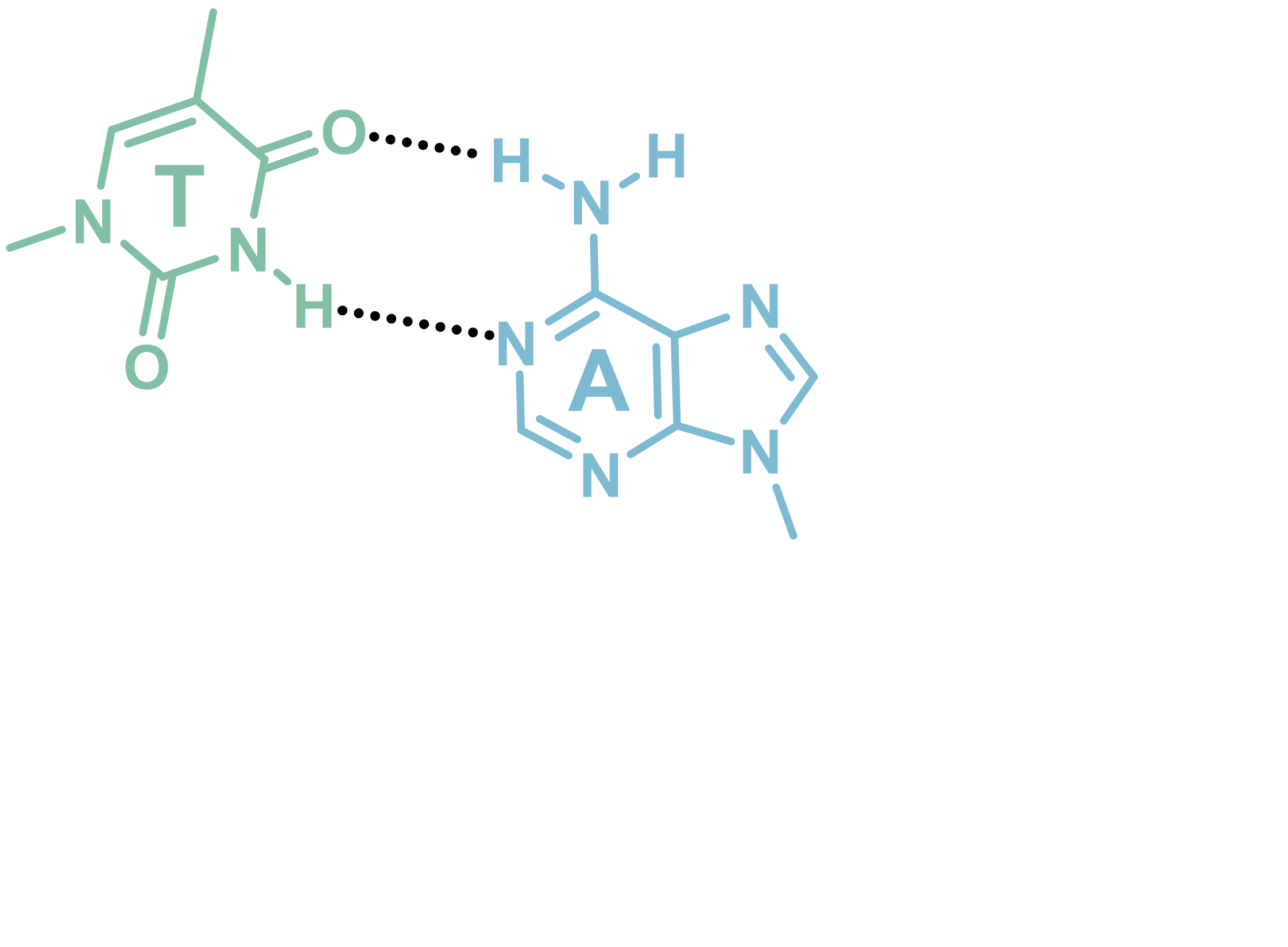
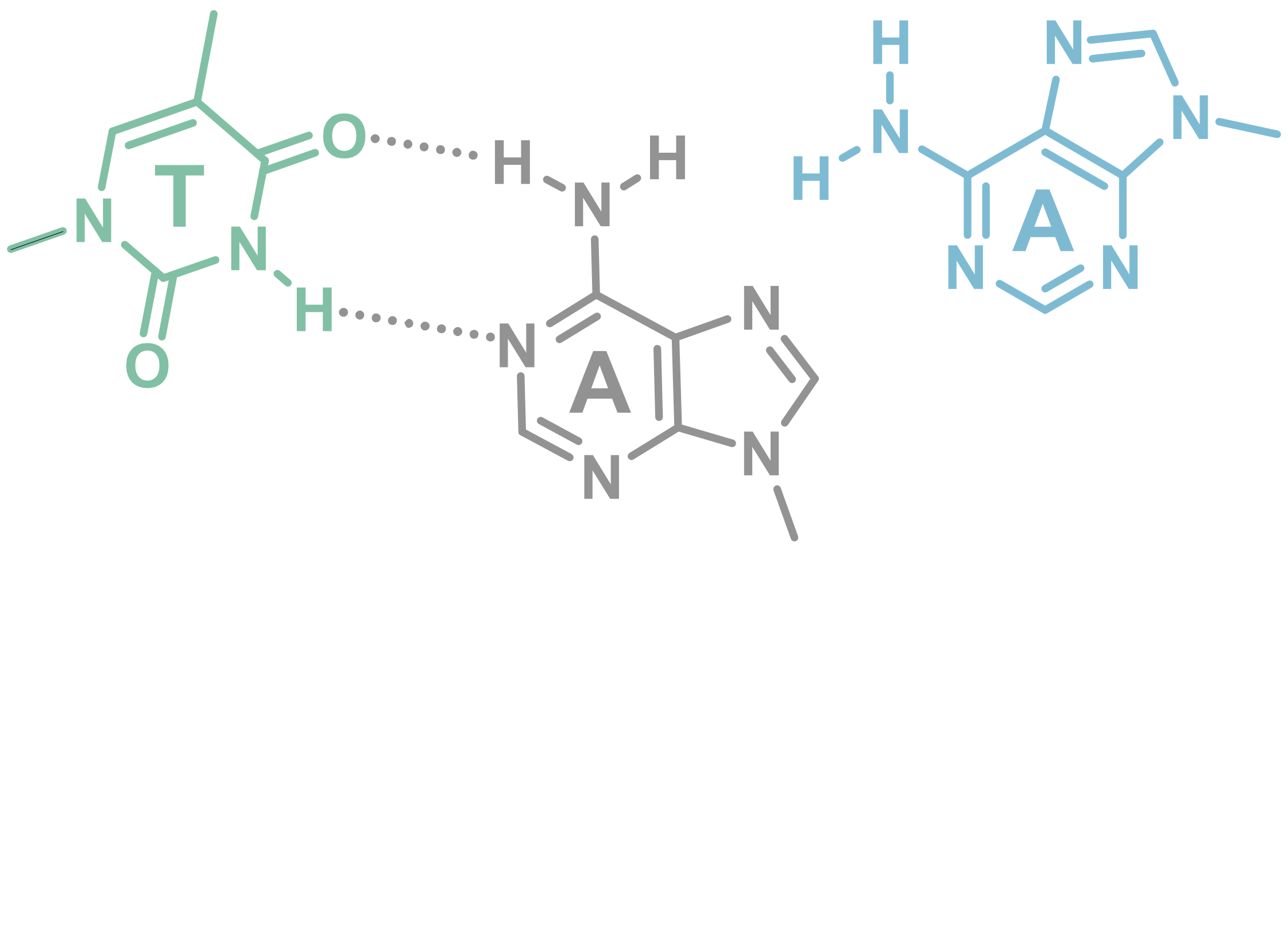
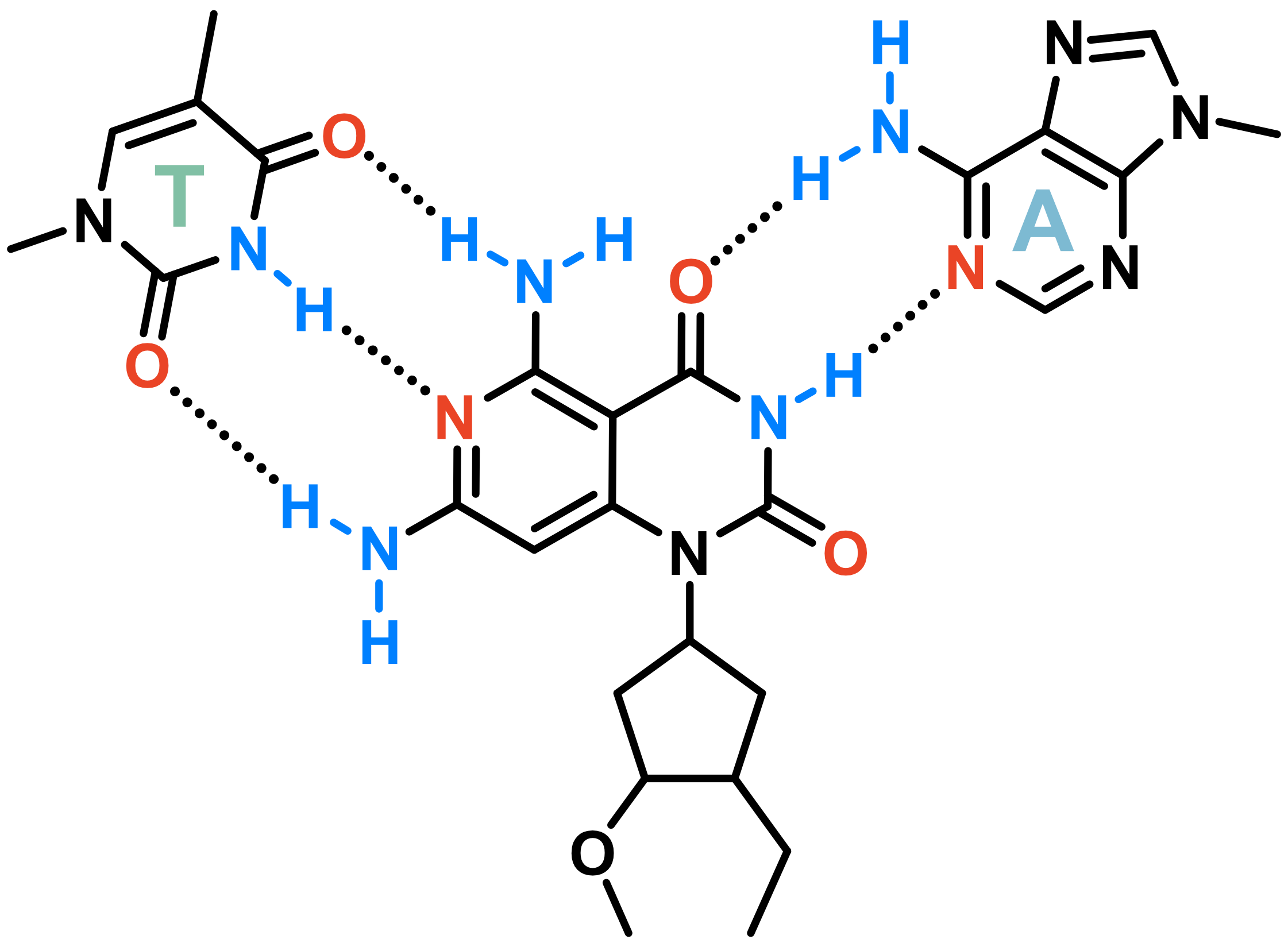
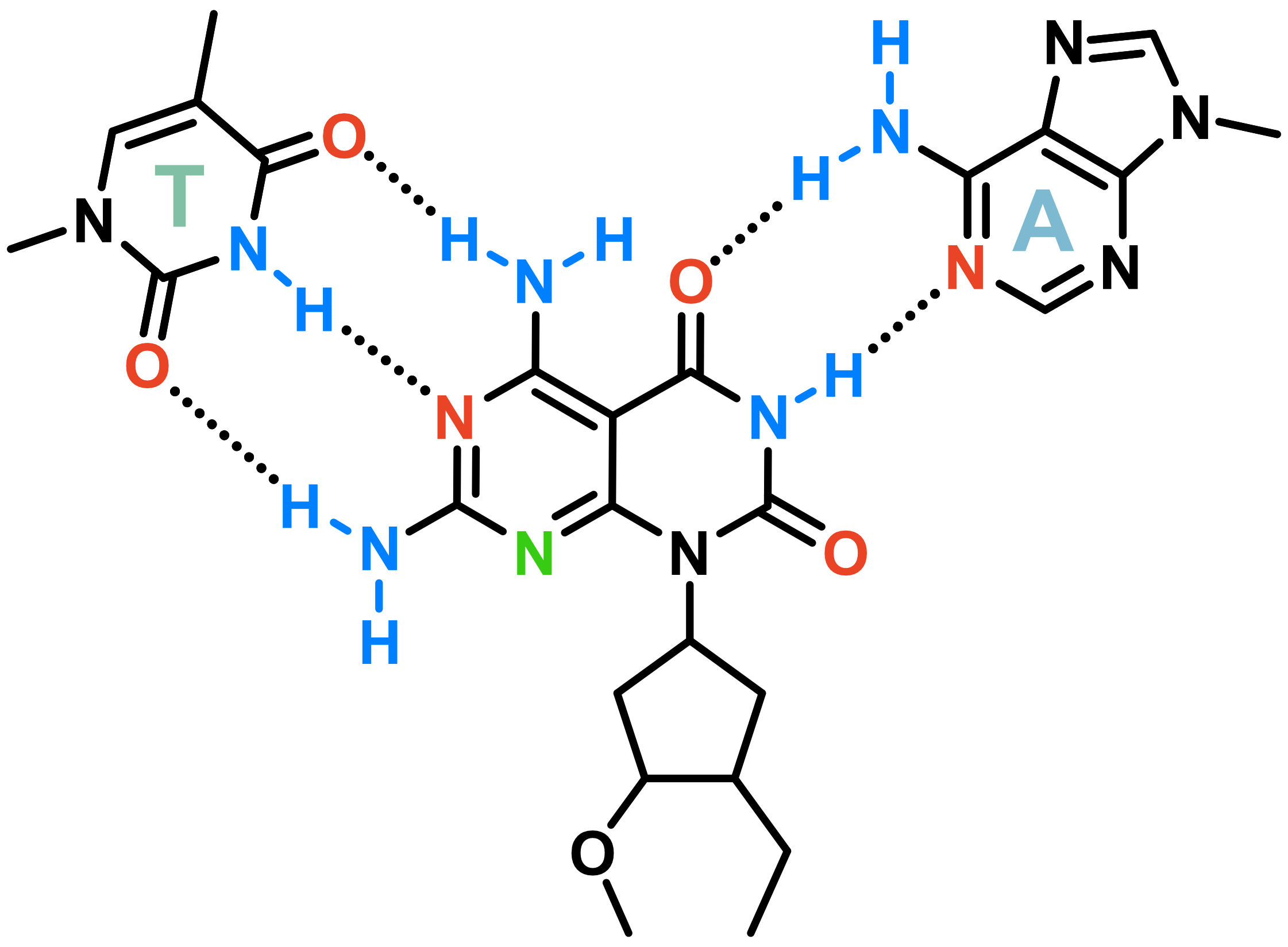

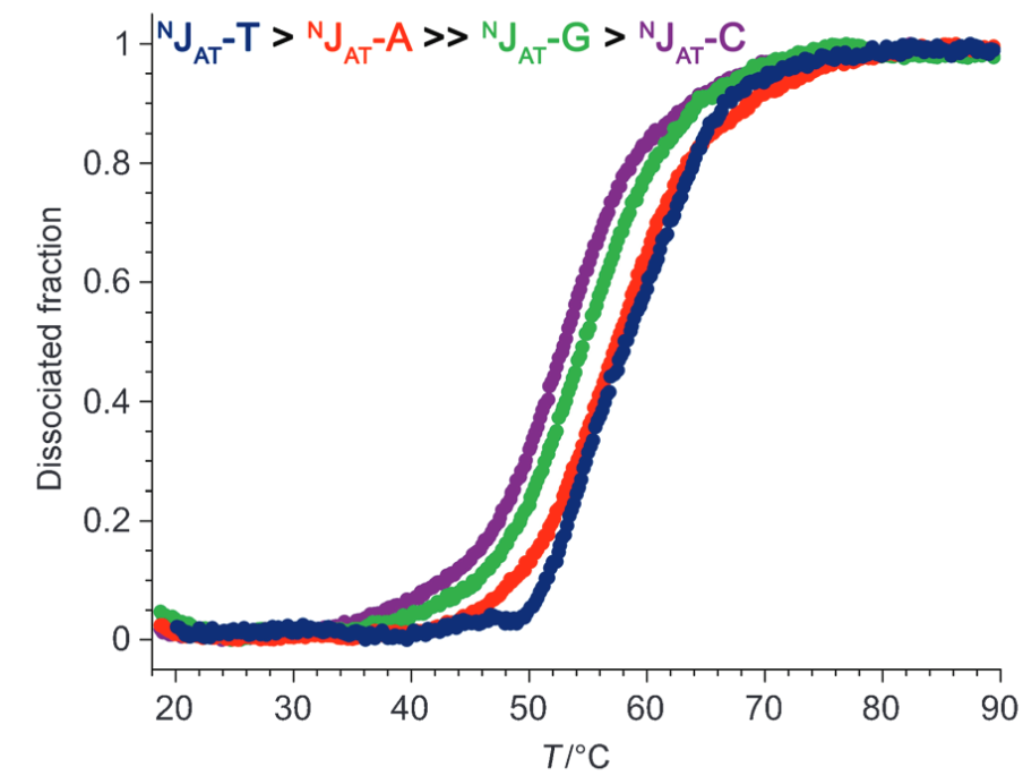
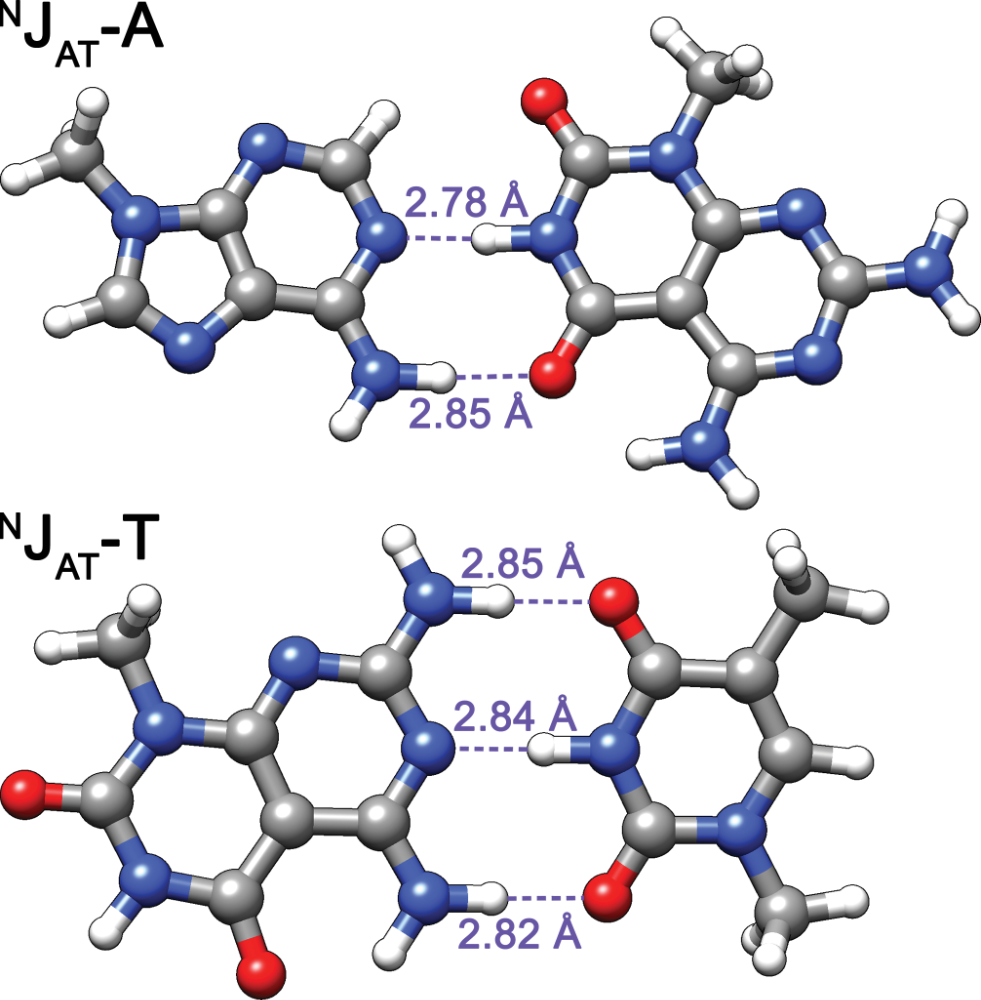
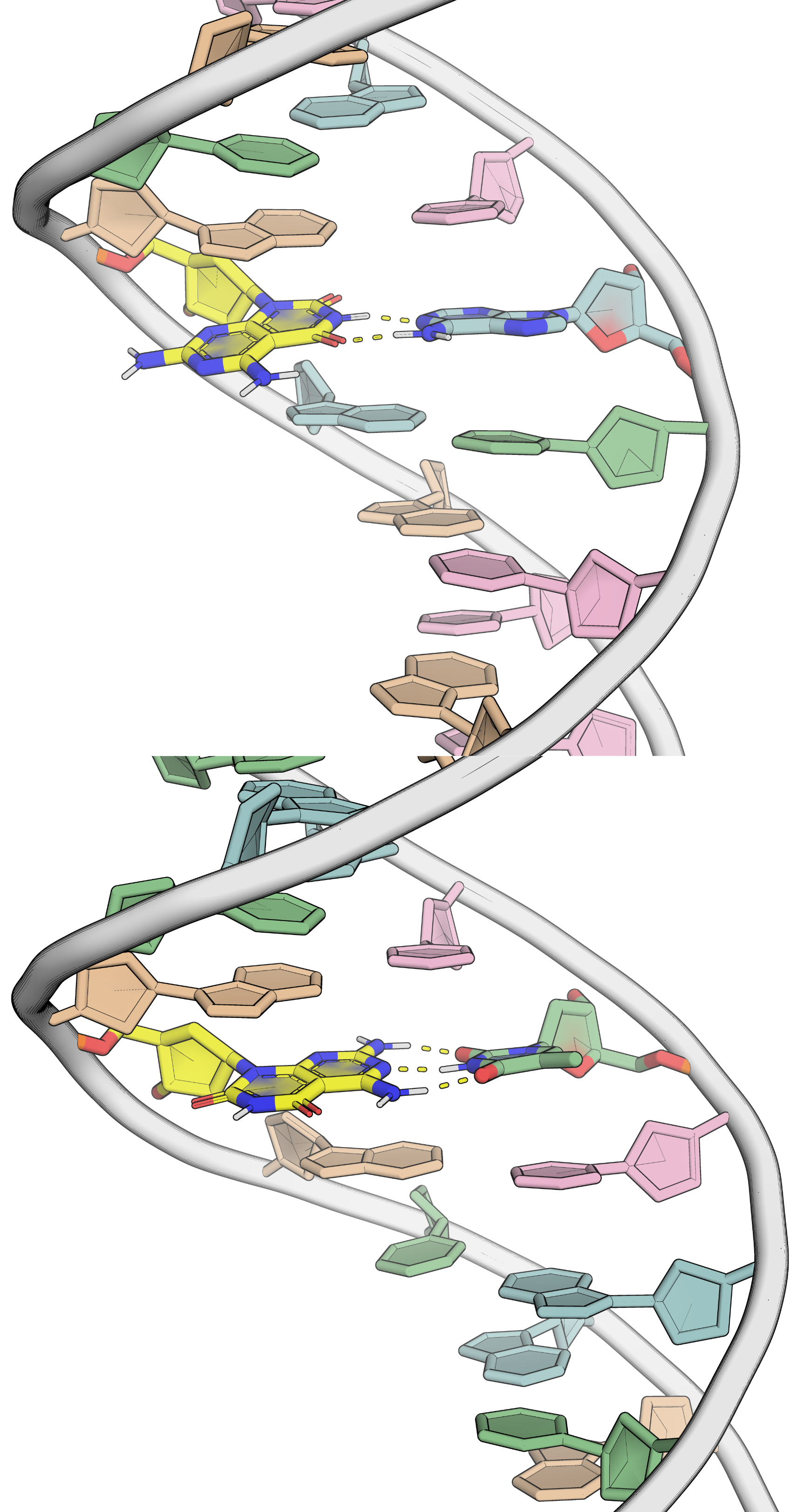

Largy, E., Liu, W., Hasan, A. et Perrin, D.M. ChemBioChem, 2013, 14, 2199; Largy, E., Liu, W., Hasan, A. et Perrin, D.M. Chem. Eur. J., 2014, 20, 1495.
Postdoc at IECB

Size-exclusion HPLC to study nucleic acid polymorphism
- Goal: develop amethod to assess the folding and stoichiometry of oligonucleotides
- Versatile
- DNA/RNA, secondary structures
- Buffers
- Large concentration range
- Temperature
- Fast, simple, automatable
- Quantitative
- Versatile
- Excellent linearity
- Application on real-life RNA hairpins
- Here at 10°C (favors dimers)
- G4s display larger hydrodynamic radii distributions
- Robust stoichiometry assessment
- Real-life G4s can be messy
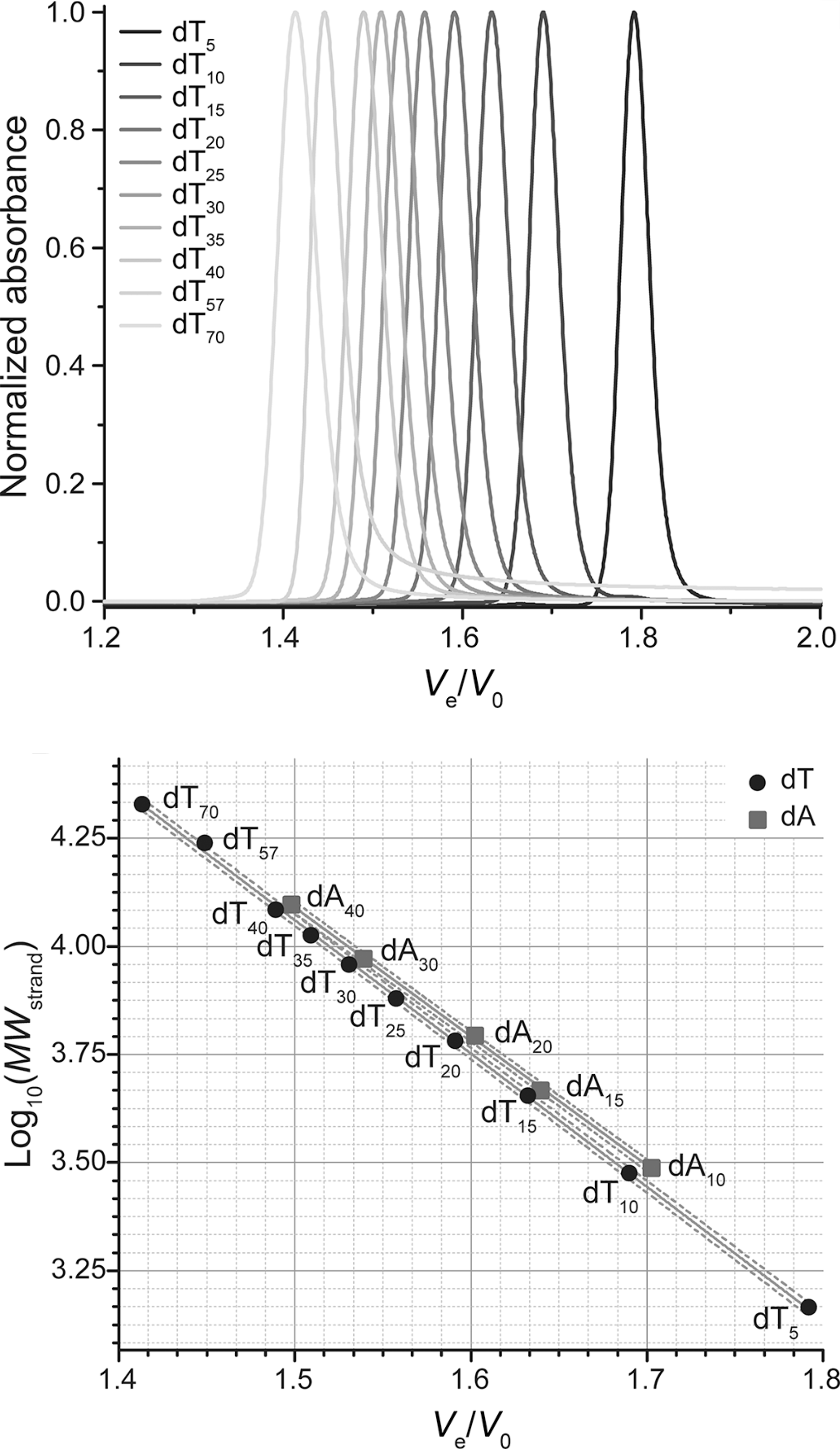
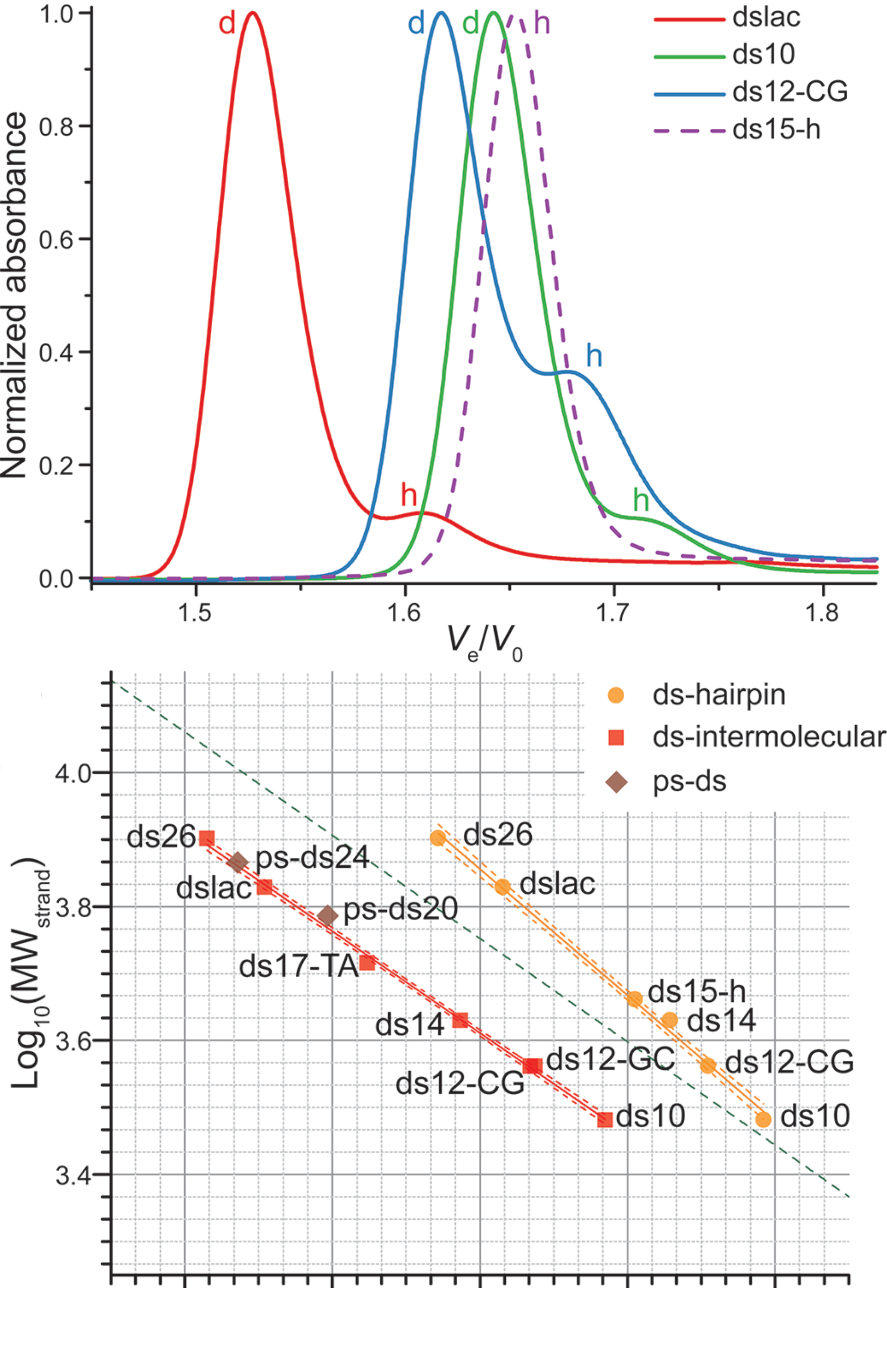
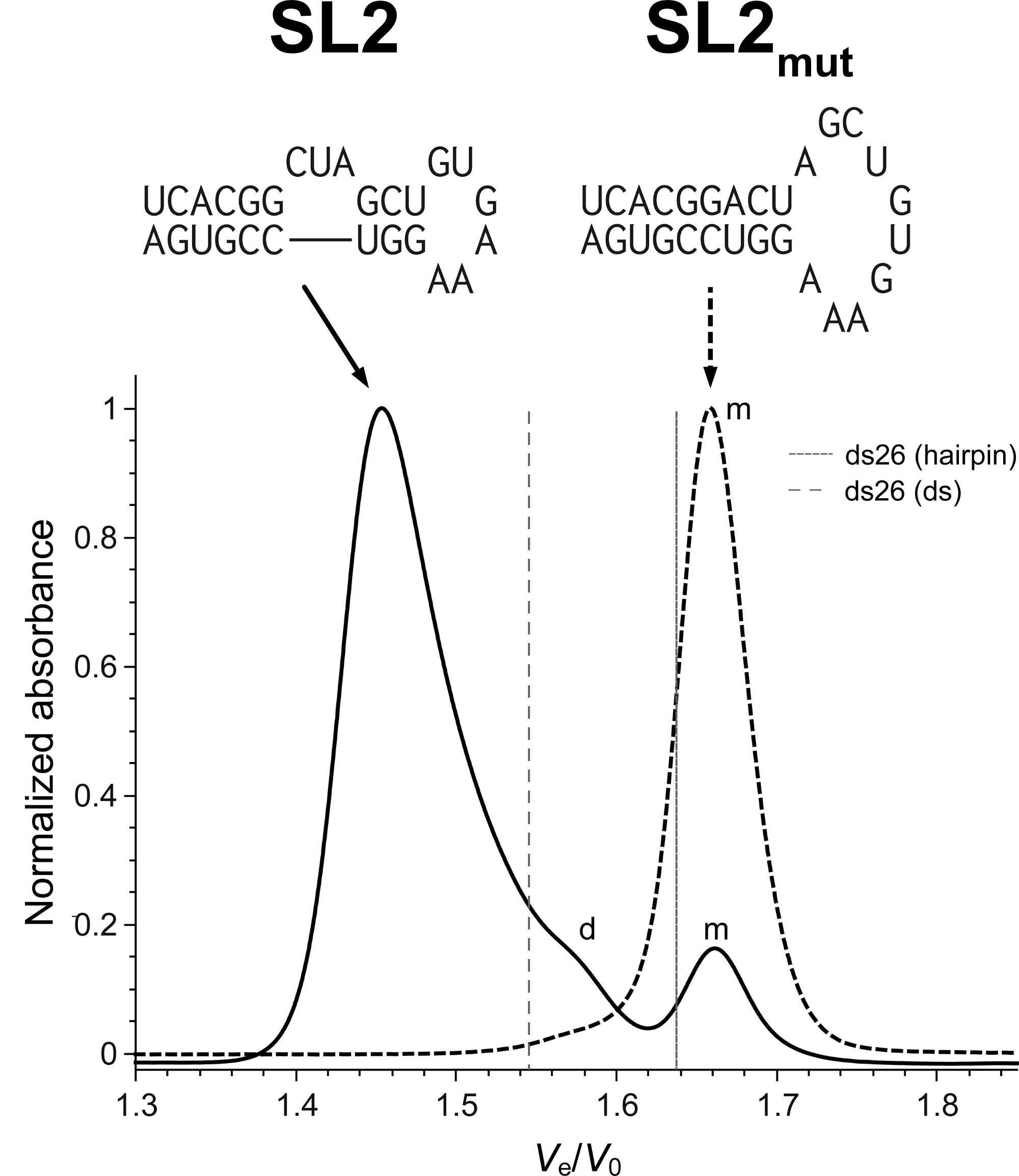
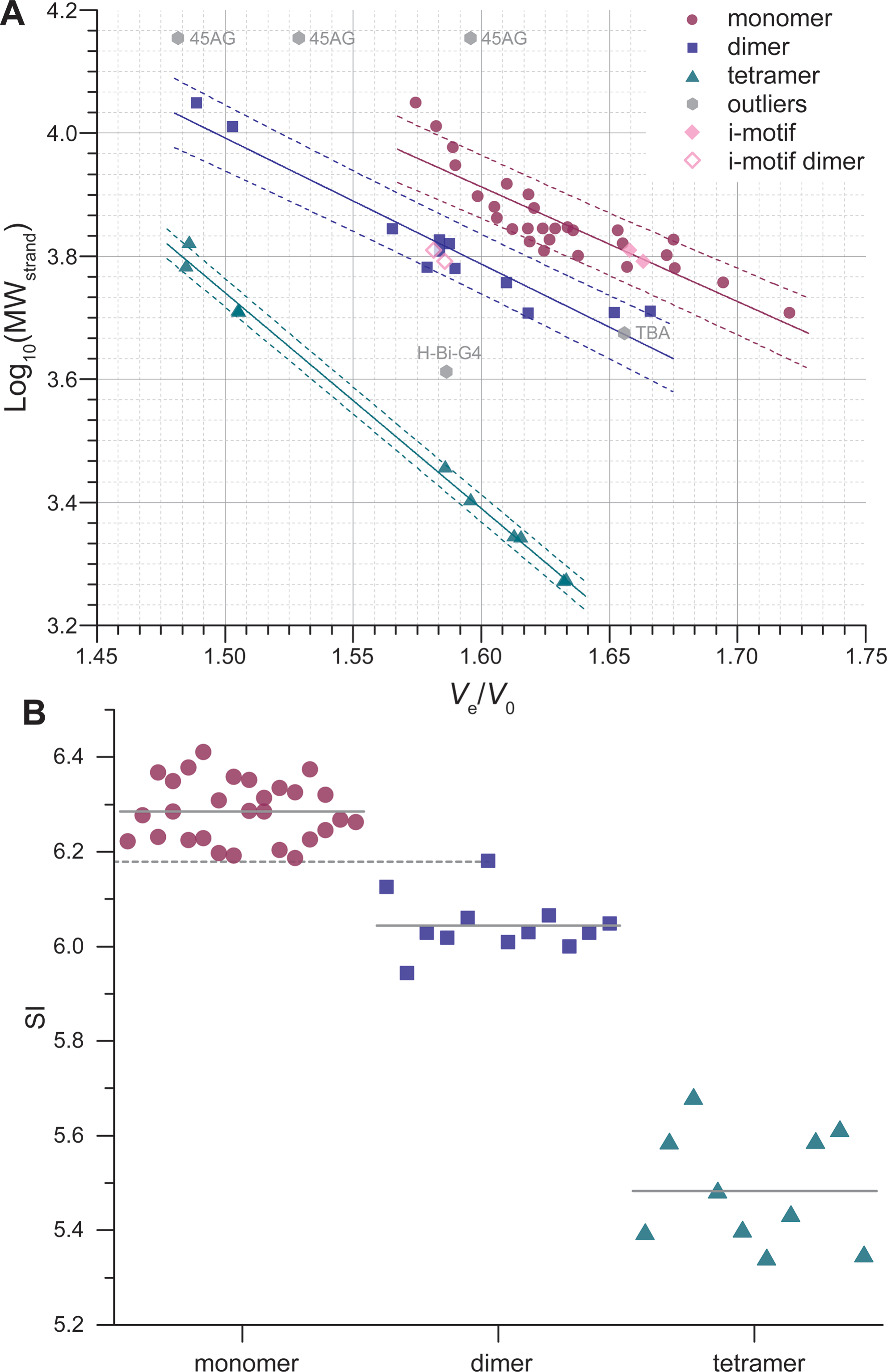
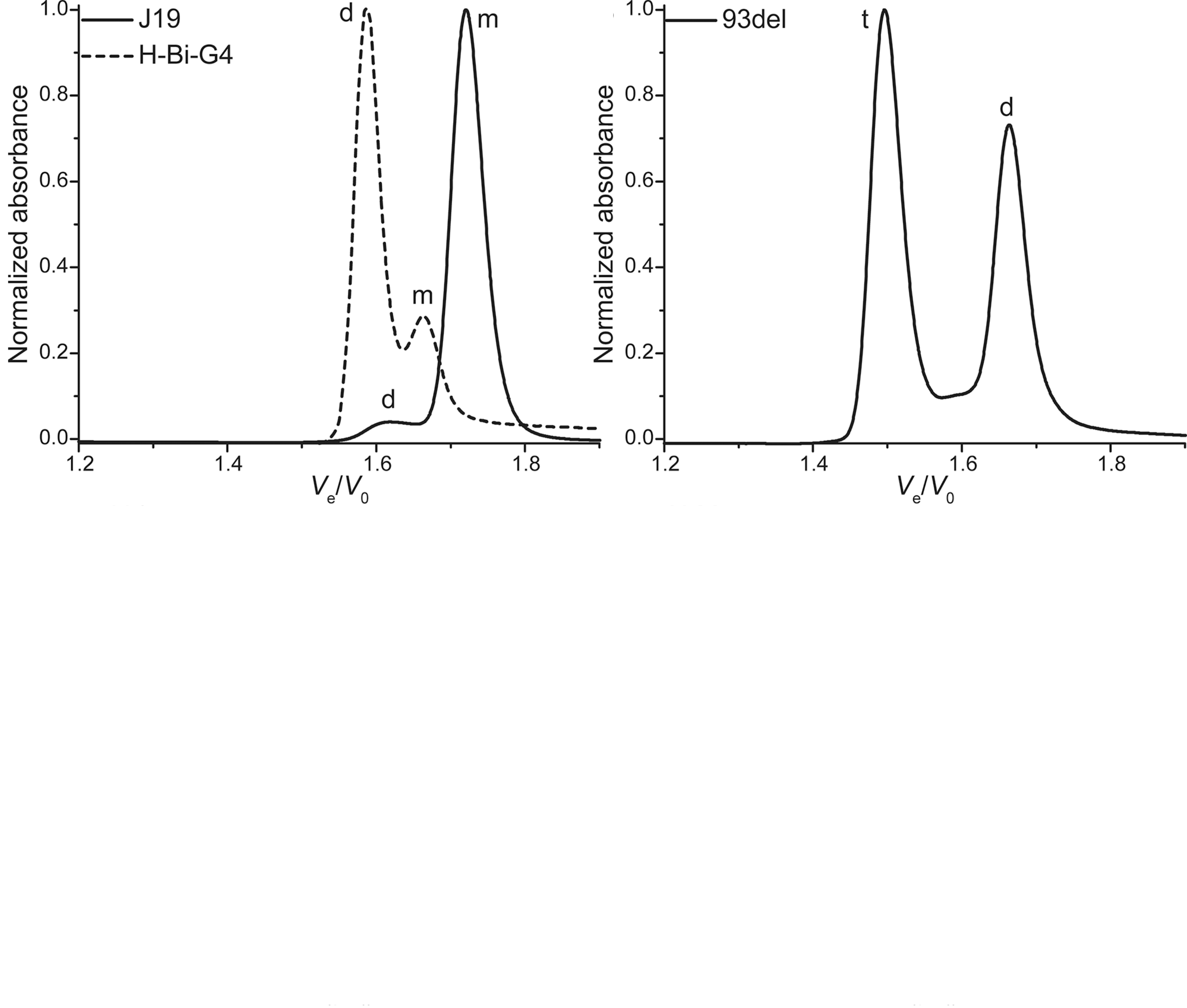
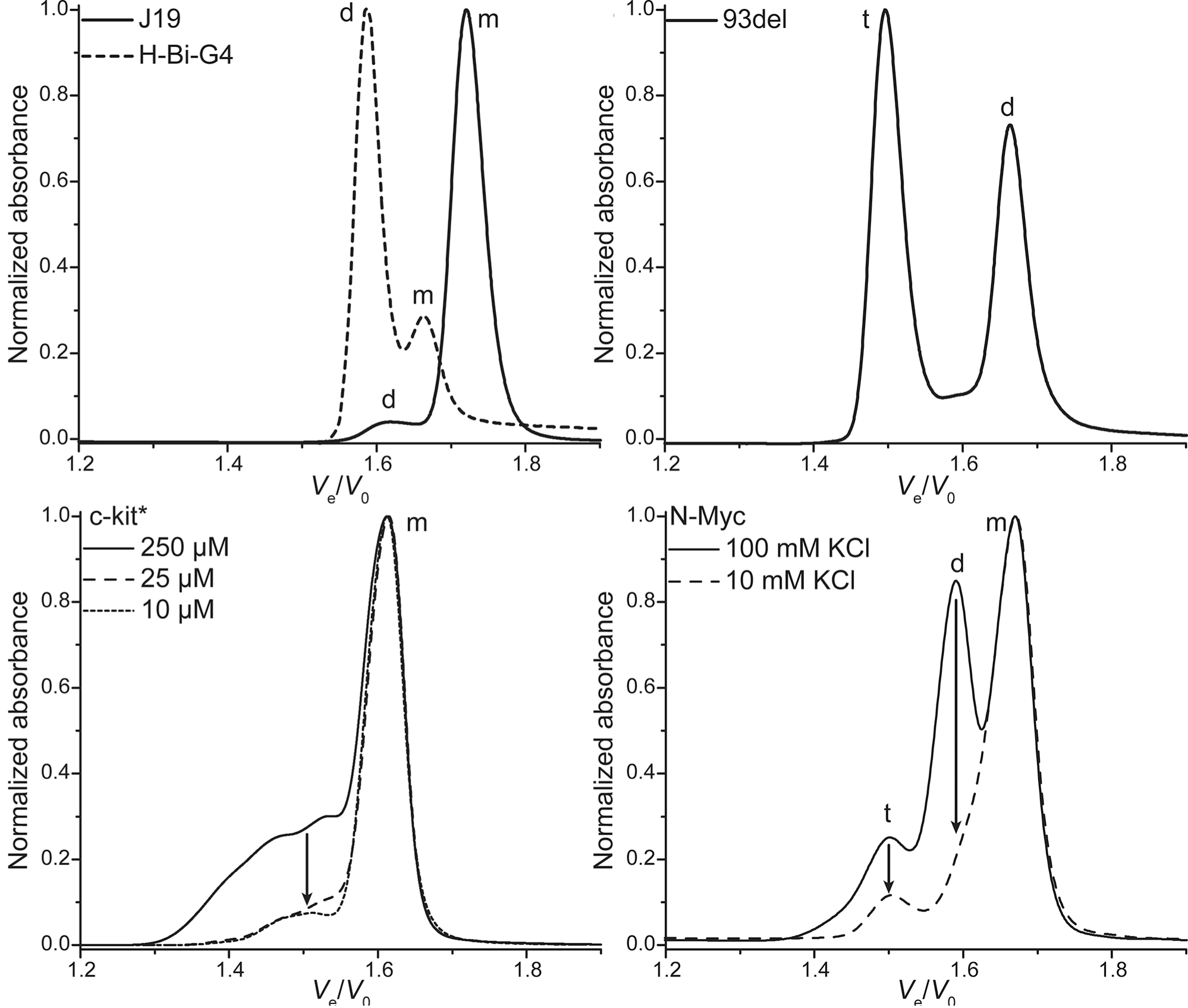
Largy, E. et Mergny, J.-L. Nucleic Acids Res., 2014, 42, e149.

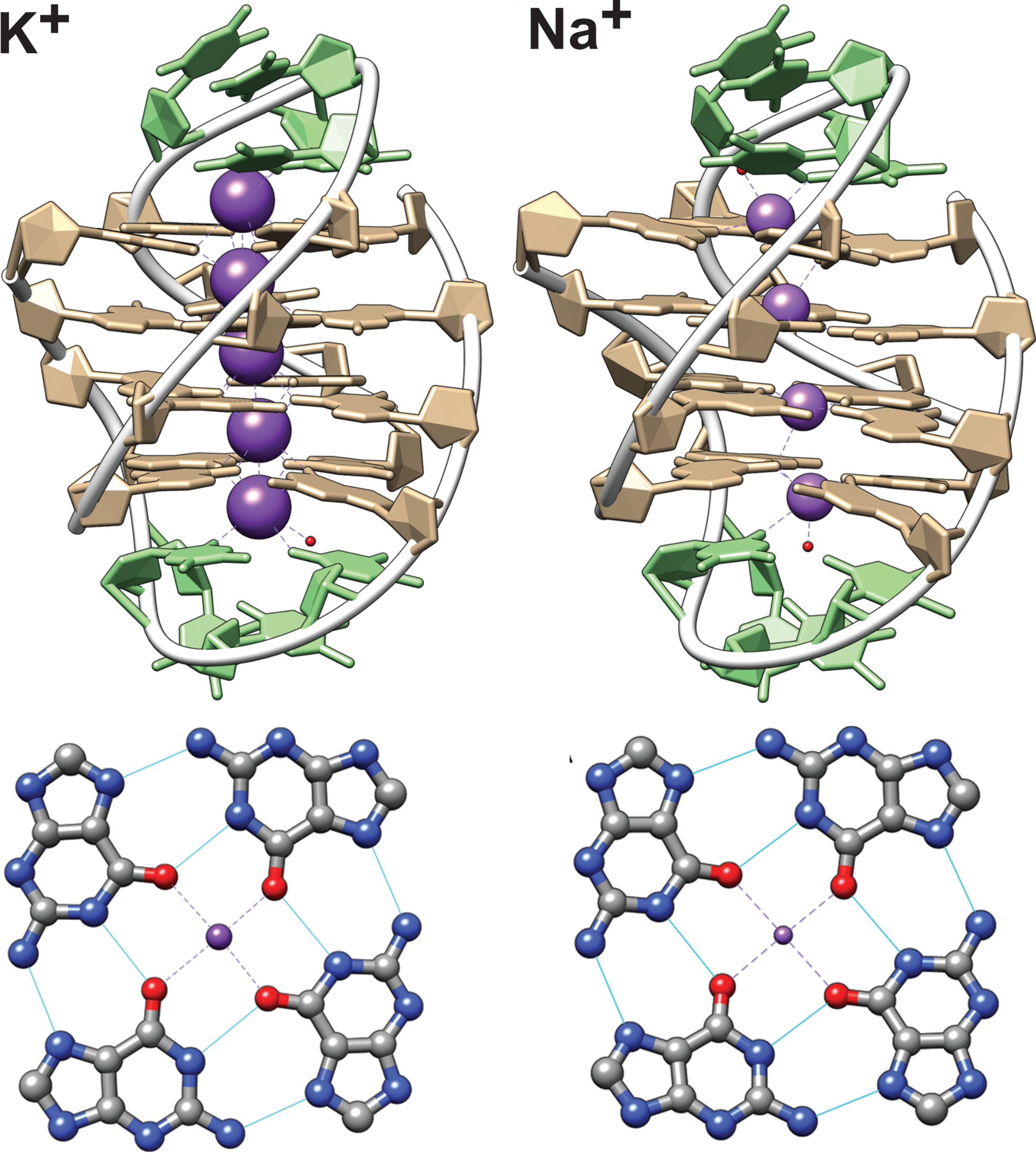
Influence of cations on G4 folding
- Cation-dependent G4 switches?
- Simple sequences!
- 222T: TGGGTTGGGTTGGGTTGGGT
- Terminal T to prevent end-stacking
- Antiparallel-to-parallel switch at low [KCl]
- Extensive characterization
- Fast (second-scale at 20°C)
- Large \(\Delta T_{m}\) between K+ and Na+
- \(E_{a}\) of interconversion by UV-vis!
- Conformer revealed at low KCl
- Antiparallel
- 2-tetrad
- Double switch!
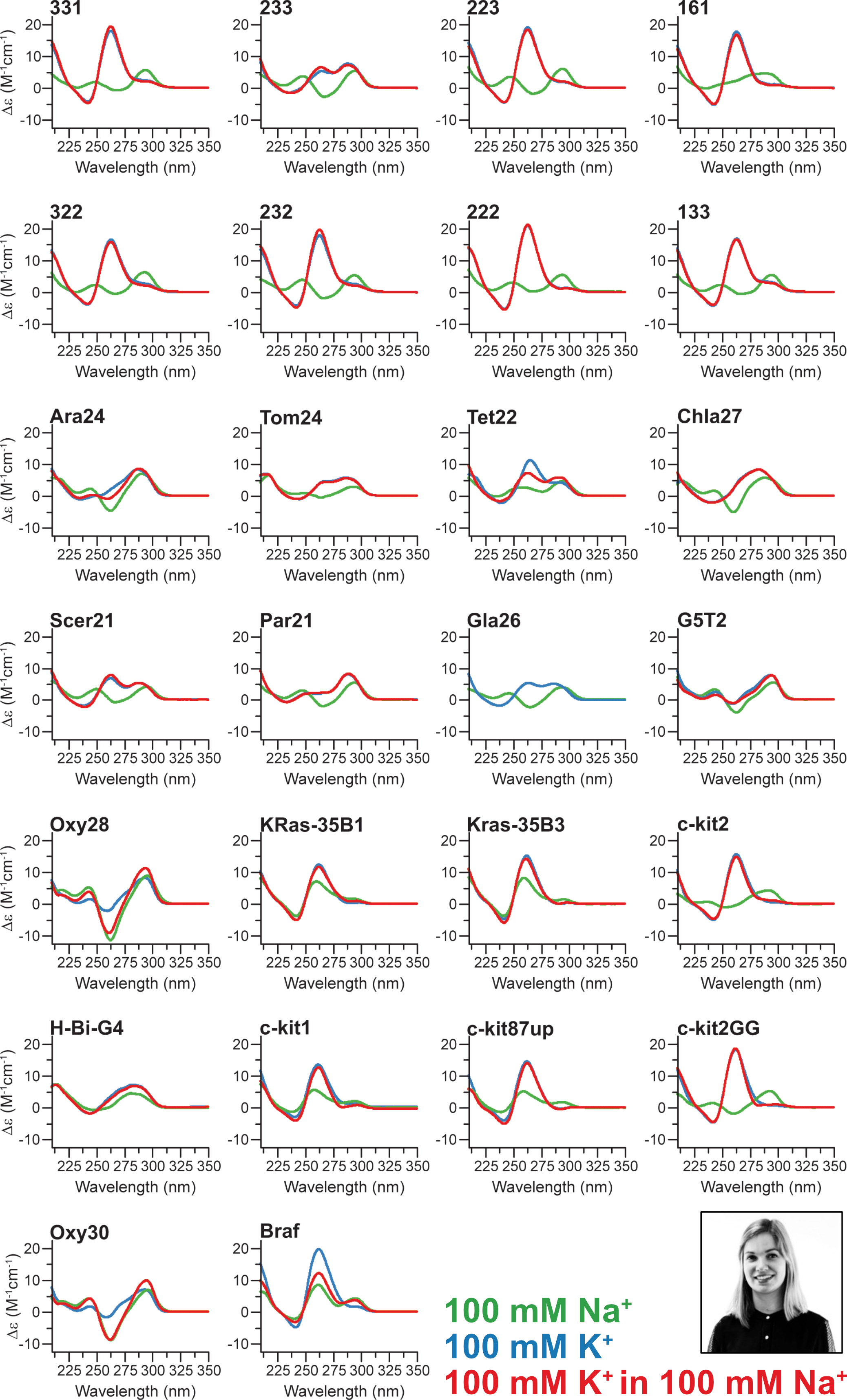
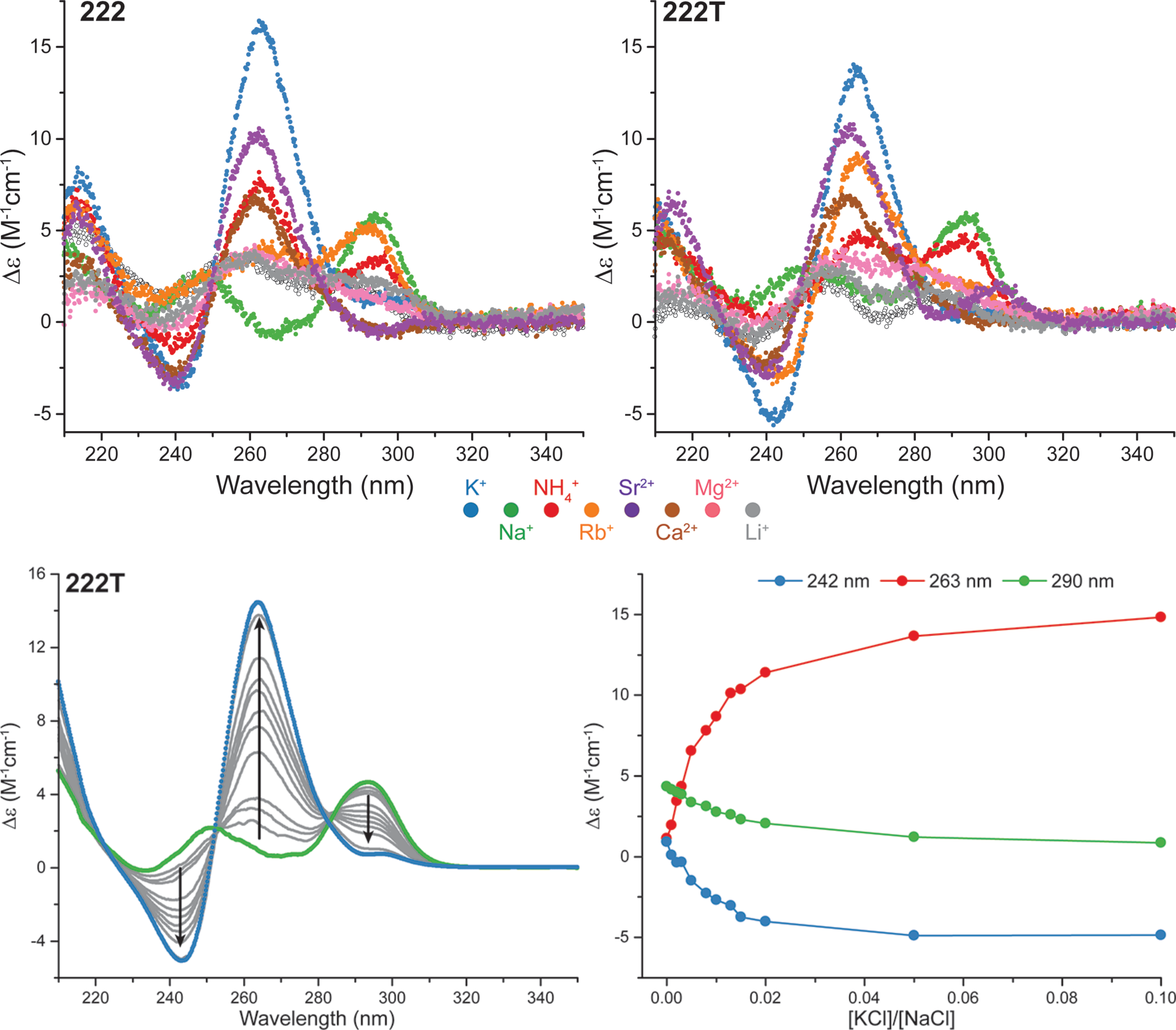
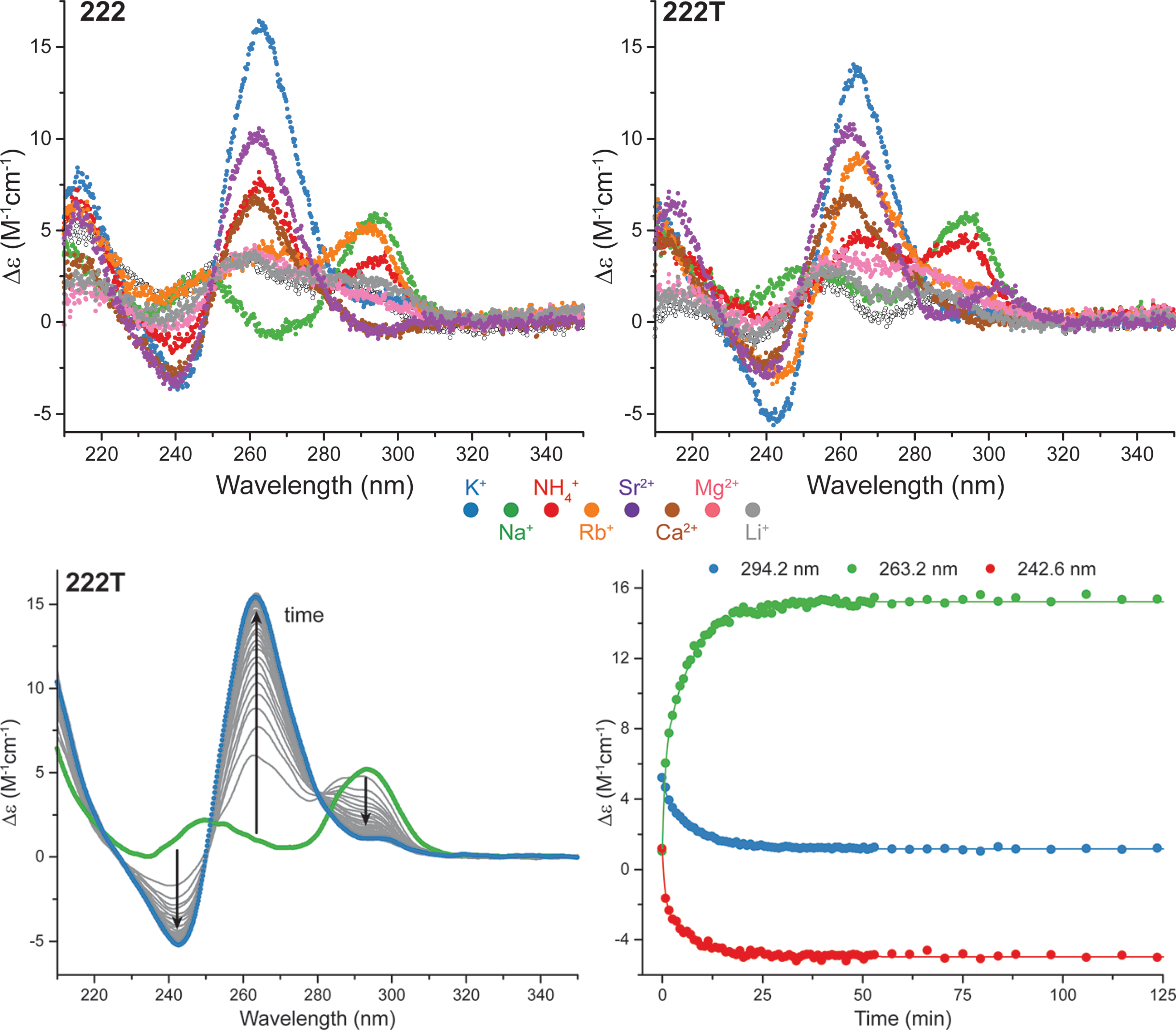
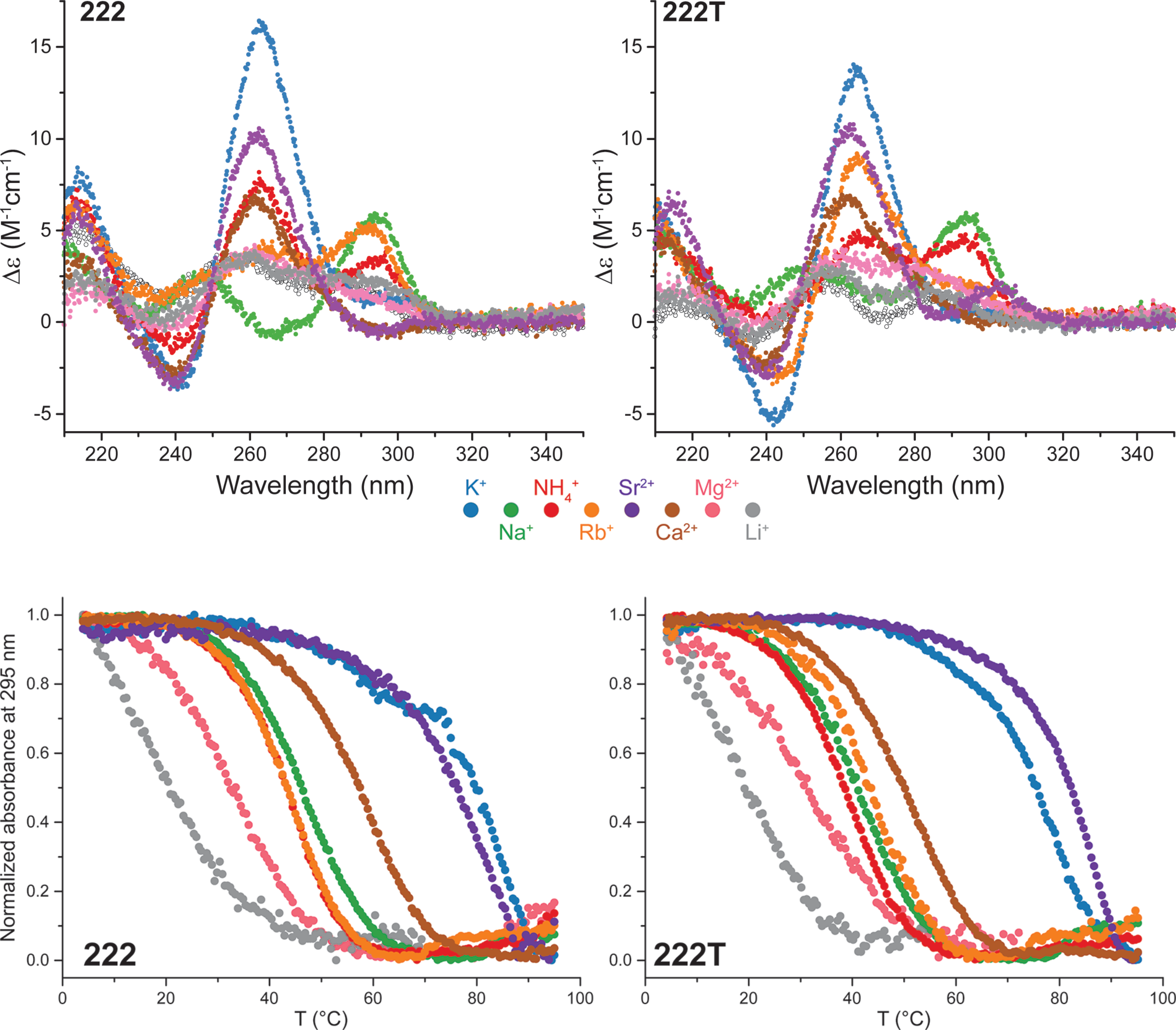

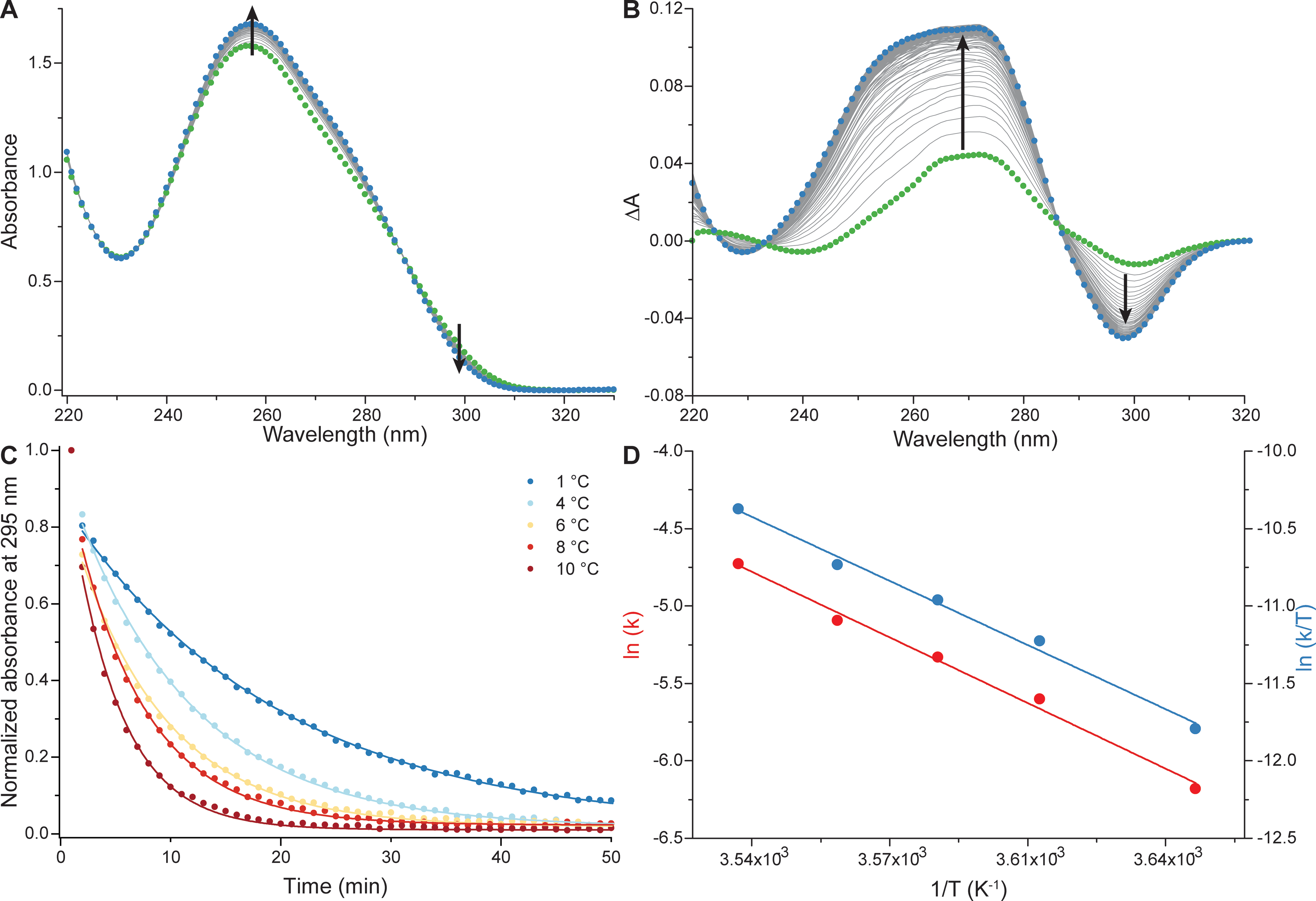
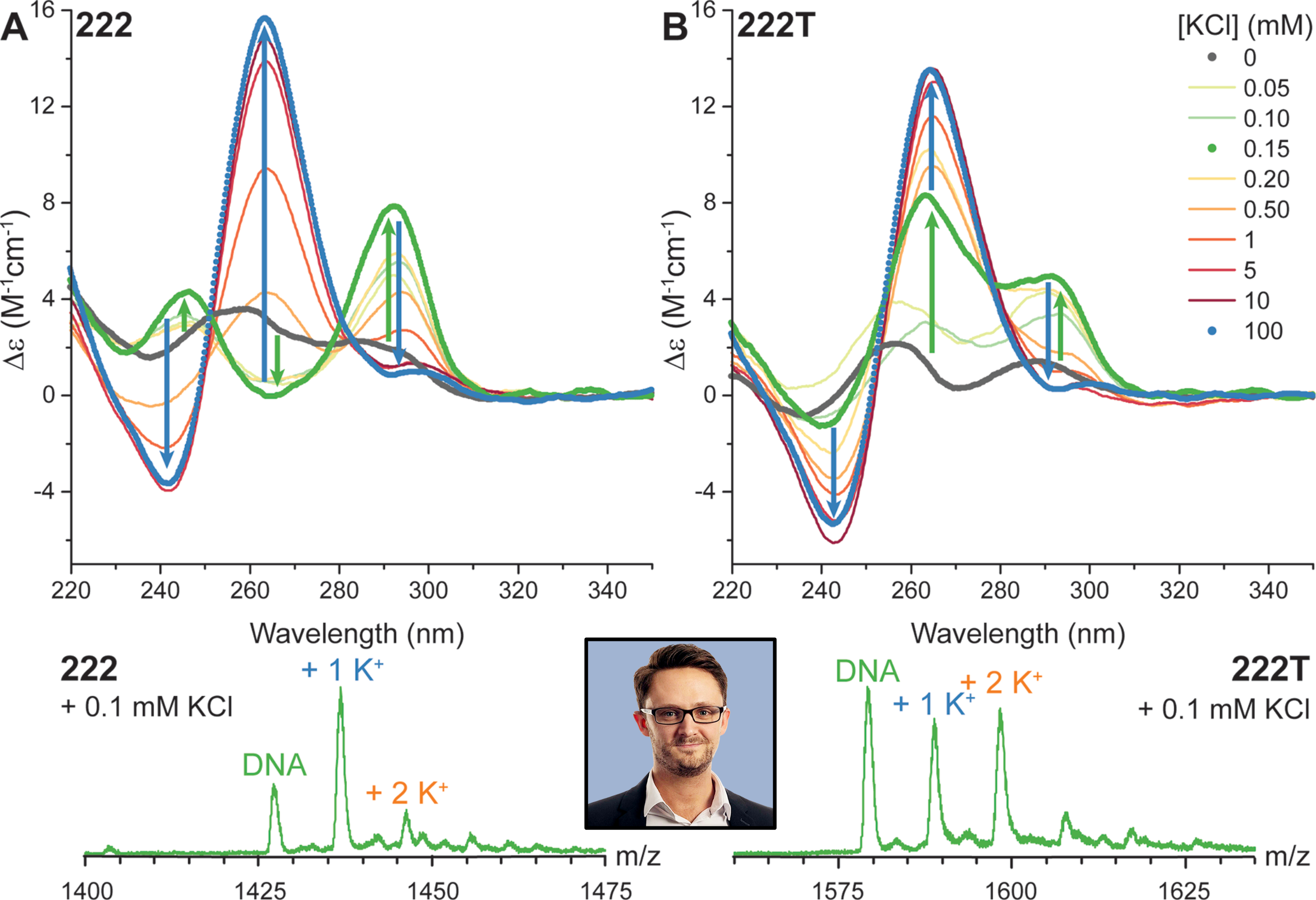

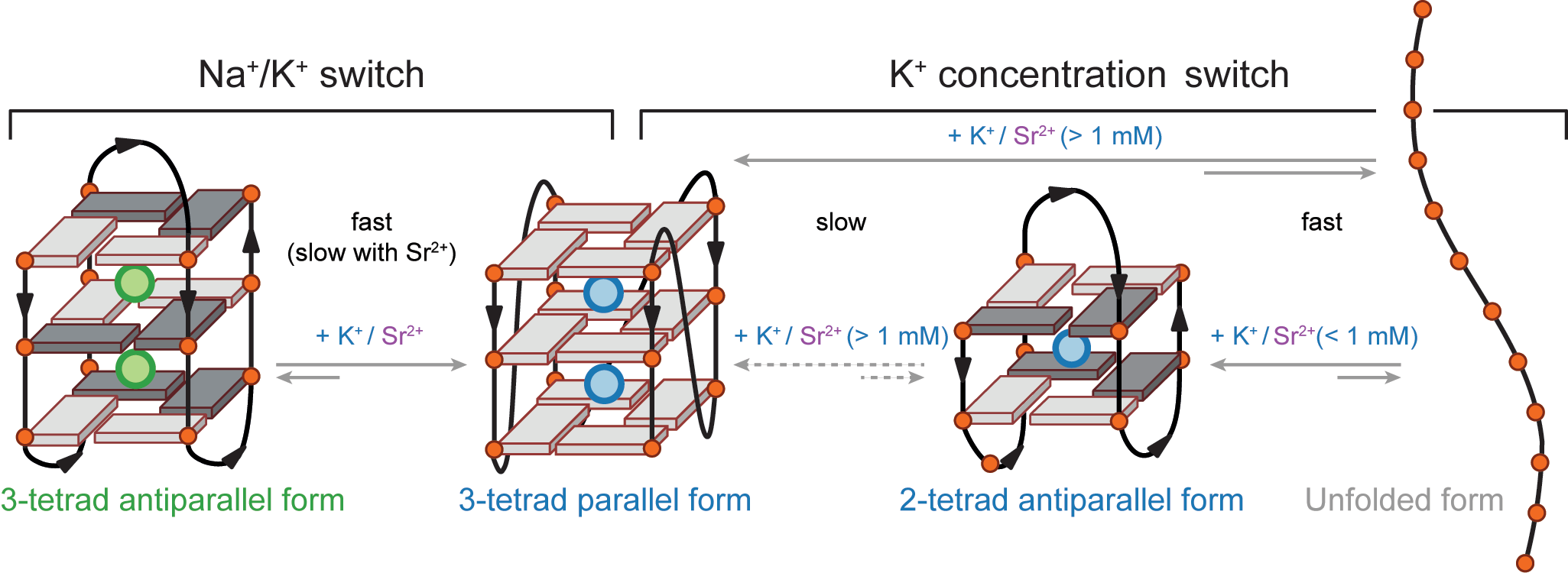


Largy, E., Marchand, A., Amrane, S., Gabelica, V. et Mergny, J.-L. J. Am. Chem. Soc., 2016, 138, 2780
Research Scientist at Quality Assistance

A quick introduction to HDX/MS
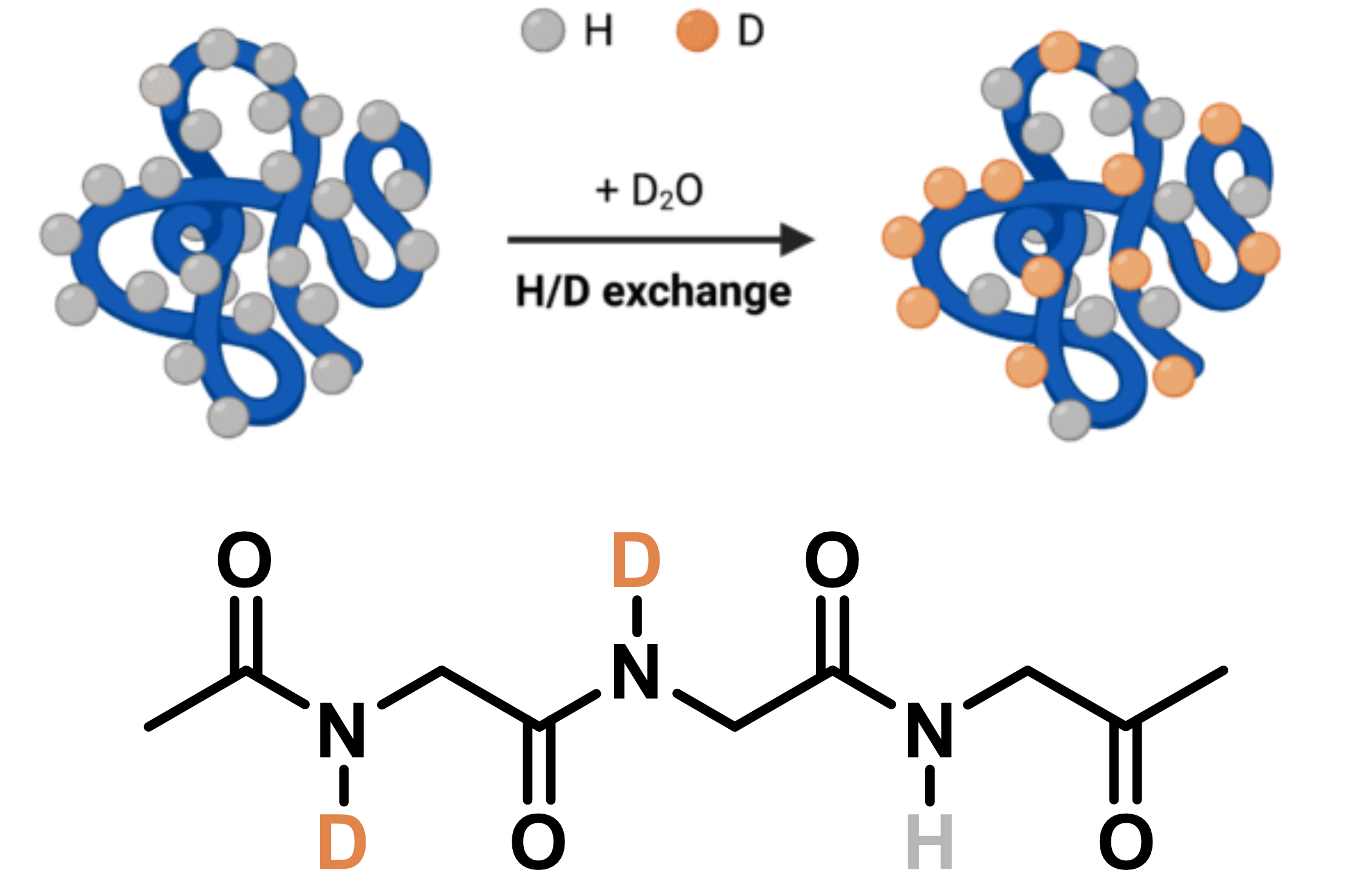
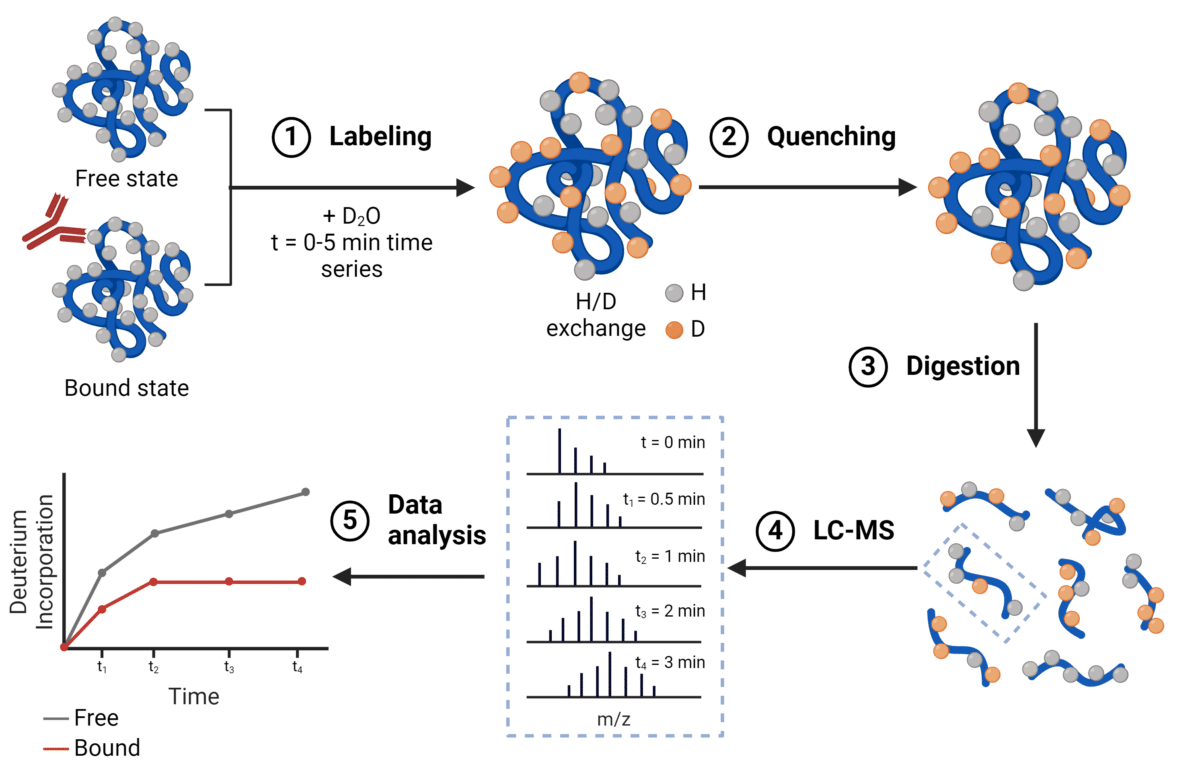

Adapted from Wang, Y. © 2024 Rapid Novor, Inc.
Epitope mapping of mAbs
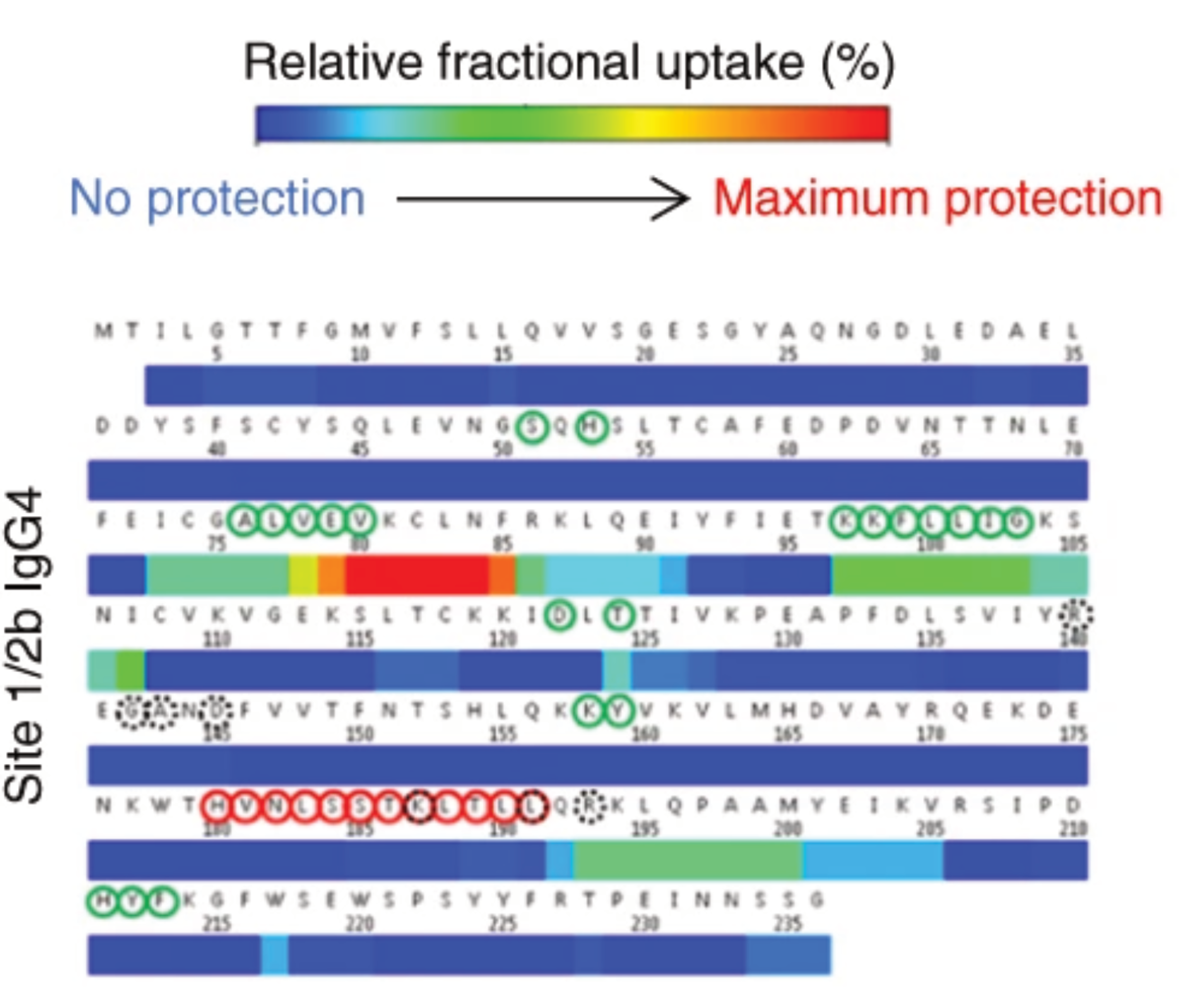
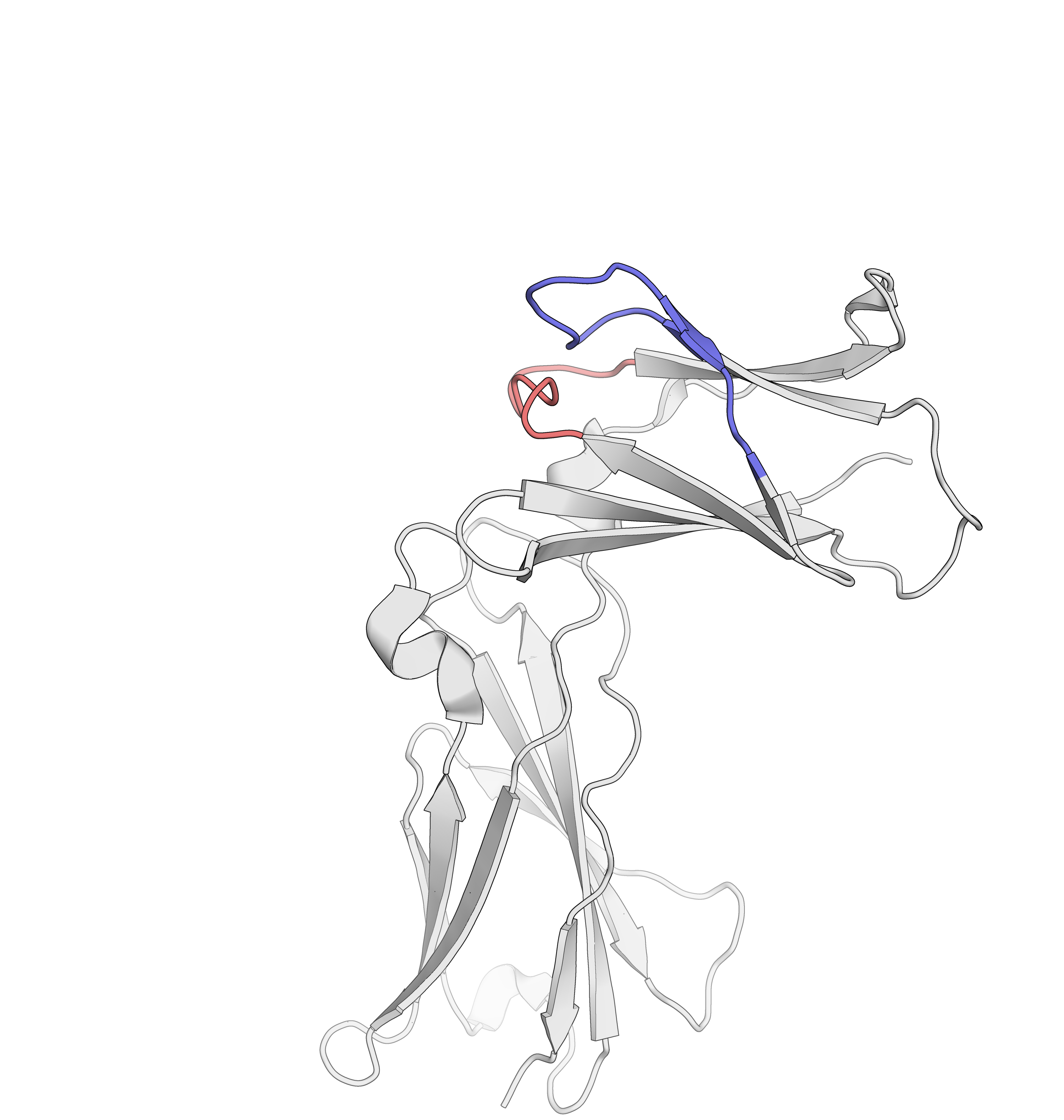
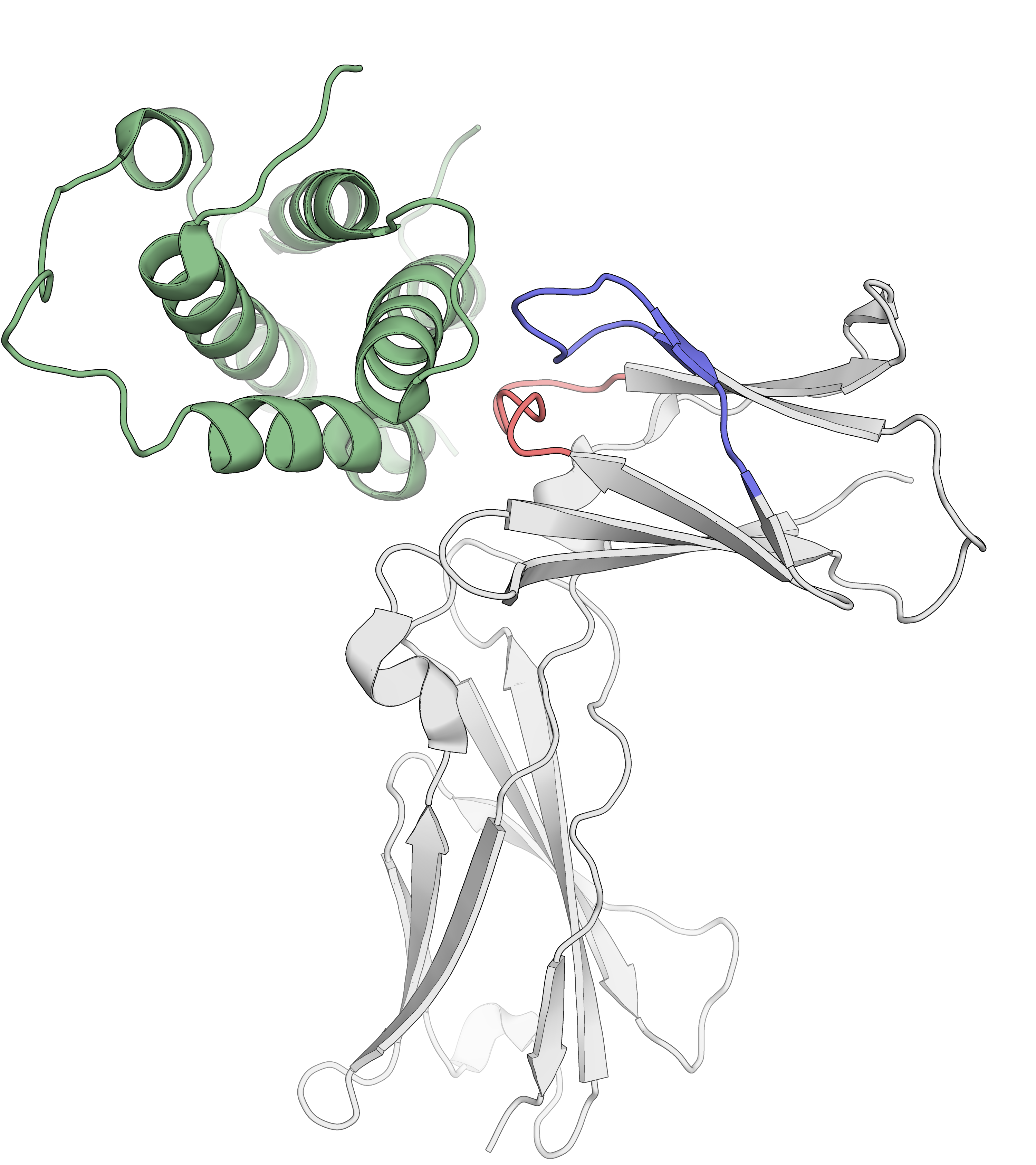
Belarif, L. et al. Nature Communications, 2018, 9, 4483
MCU at the University of Bordeaux

Native HDX/MS of nucleic acids
DNA is amenable to HDX/MS
- H/D exchange ideal for structure probing
- Full sequence coverage
- Directly involved in folding
- Minimal primary/secondary structure change
- Rate of HDX should be structure dependent
- Native MS measurements



Largy and Gabelica, Anal. Chem., 2020, 92 (6), 4402–10
Coupling in-solution HDX to native MS
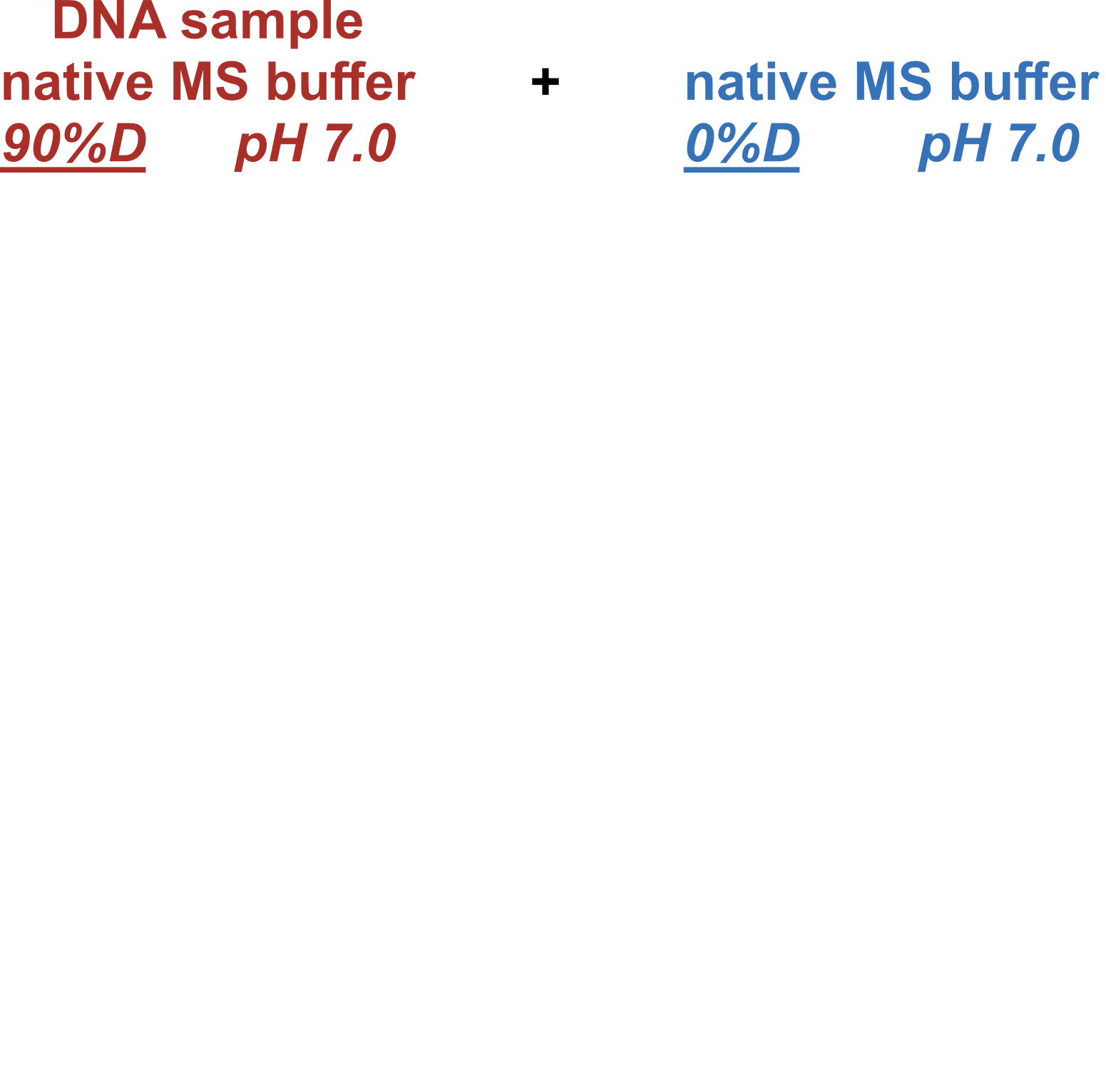
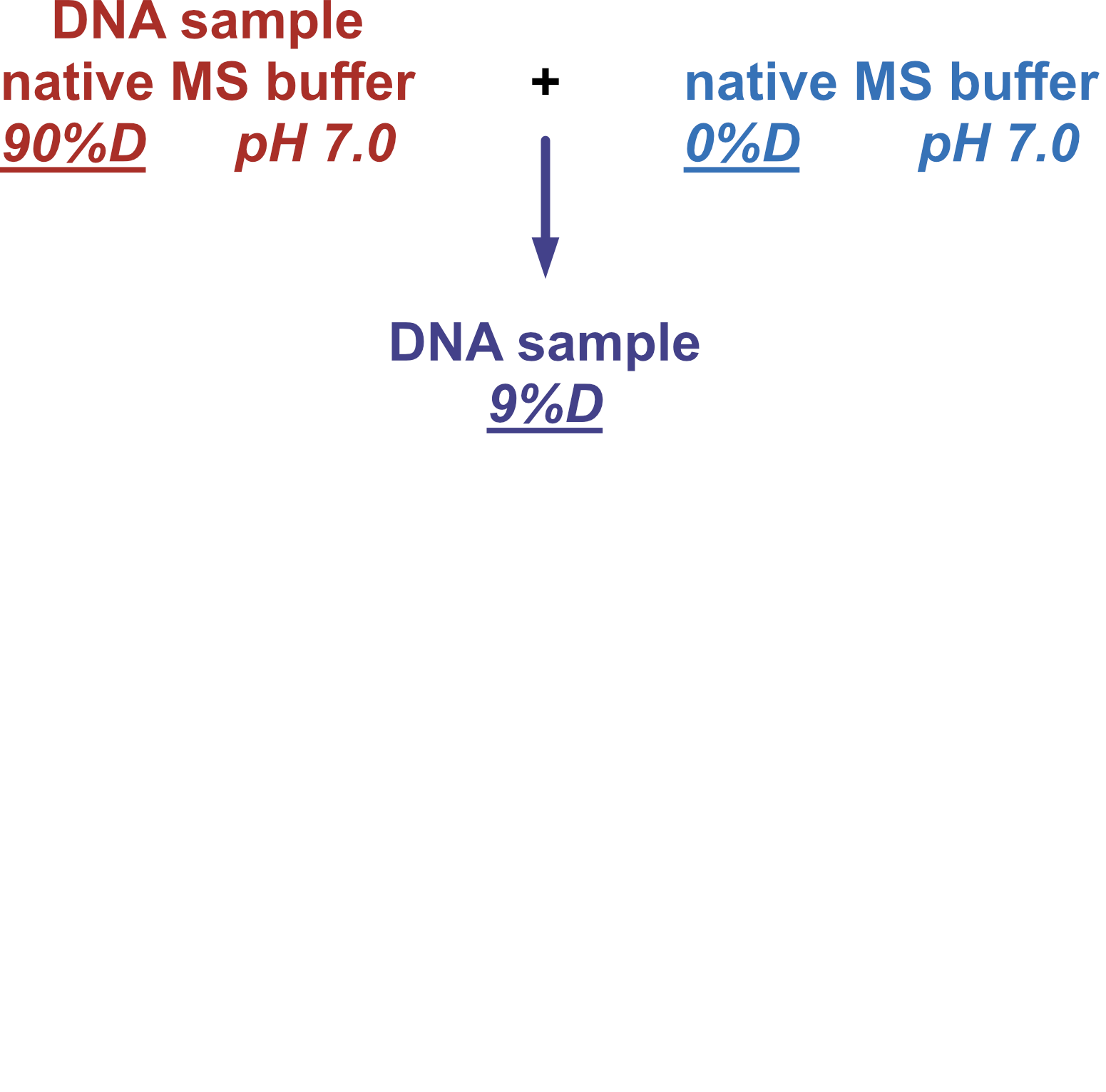
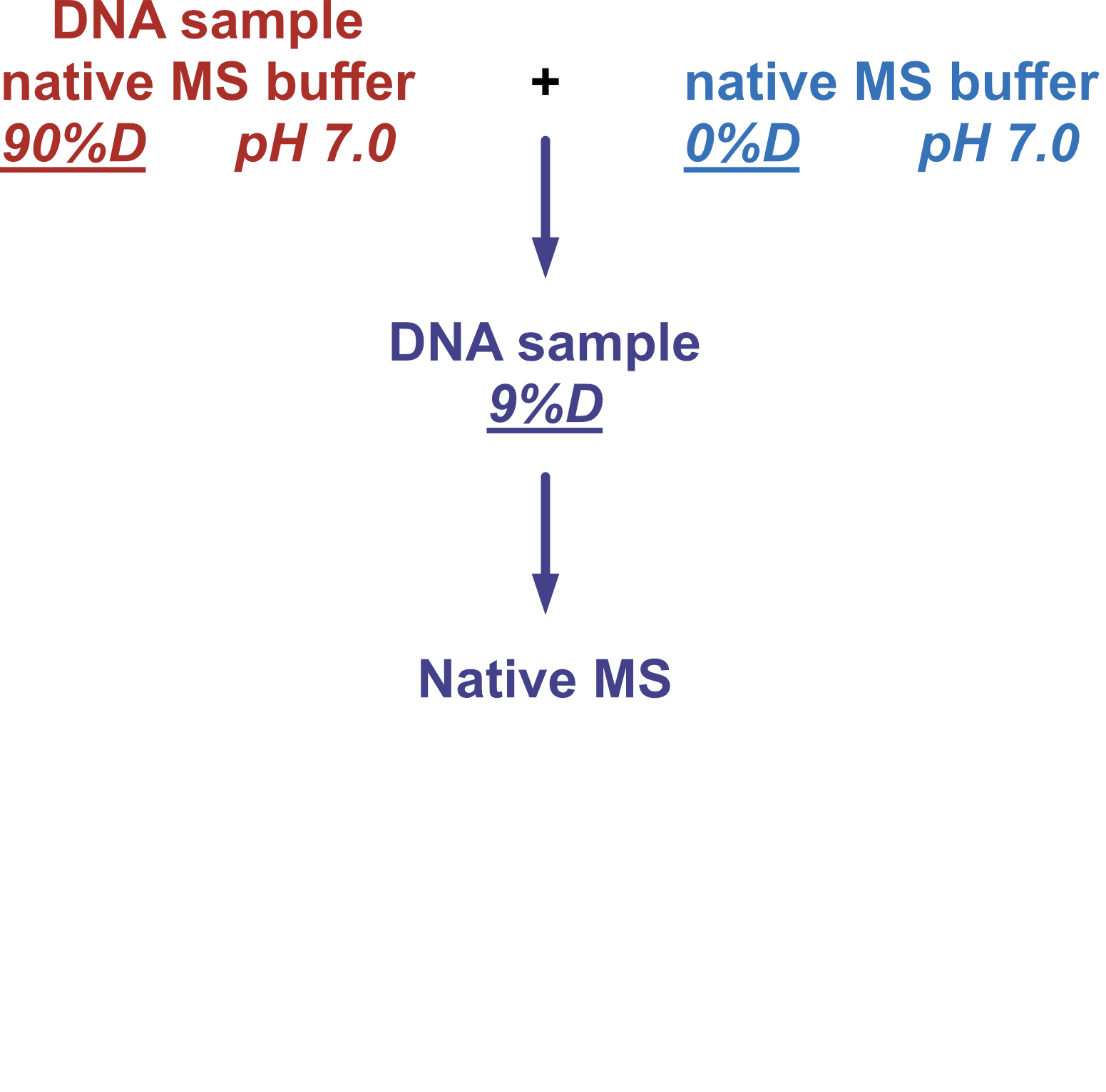



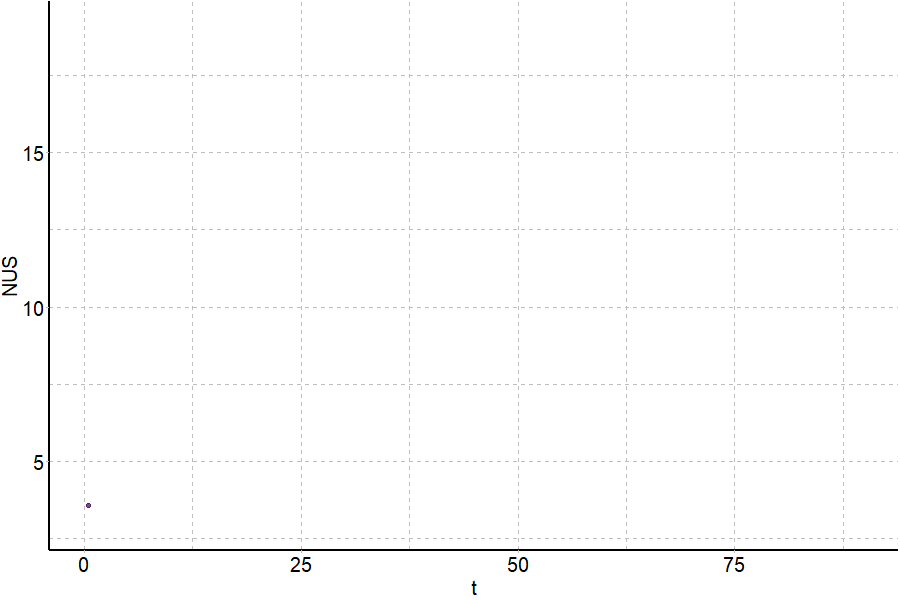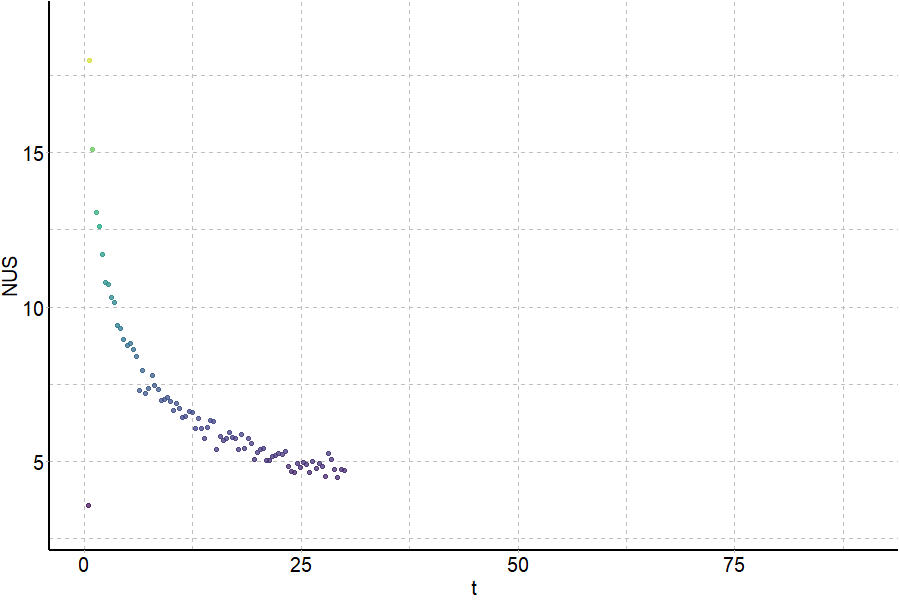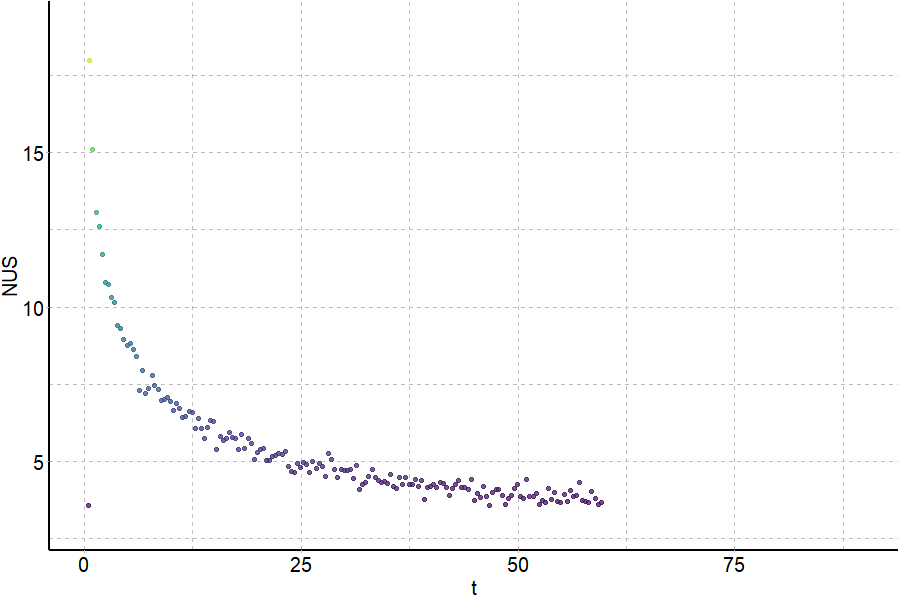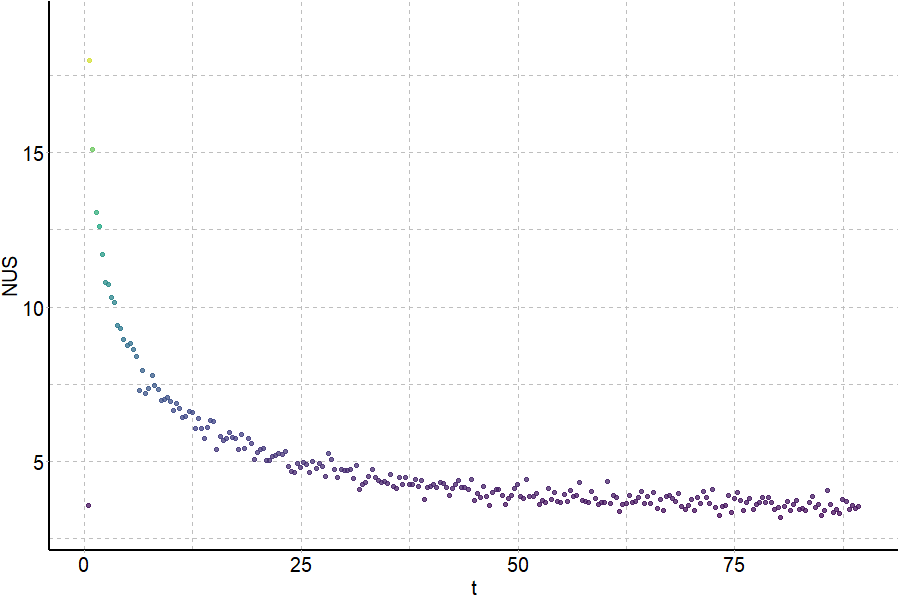
Largy and Gabelica, Anal. Chem., 2020, 92 (6), 4402–10
Largy and Ranz, Anal. Chem., 2023, 95, 9615–22
Exchange rates are sensitive to folding







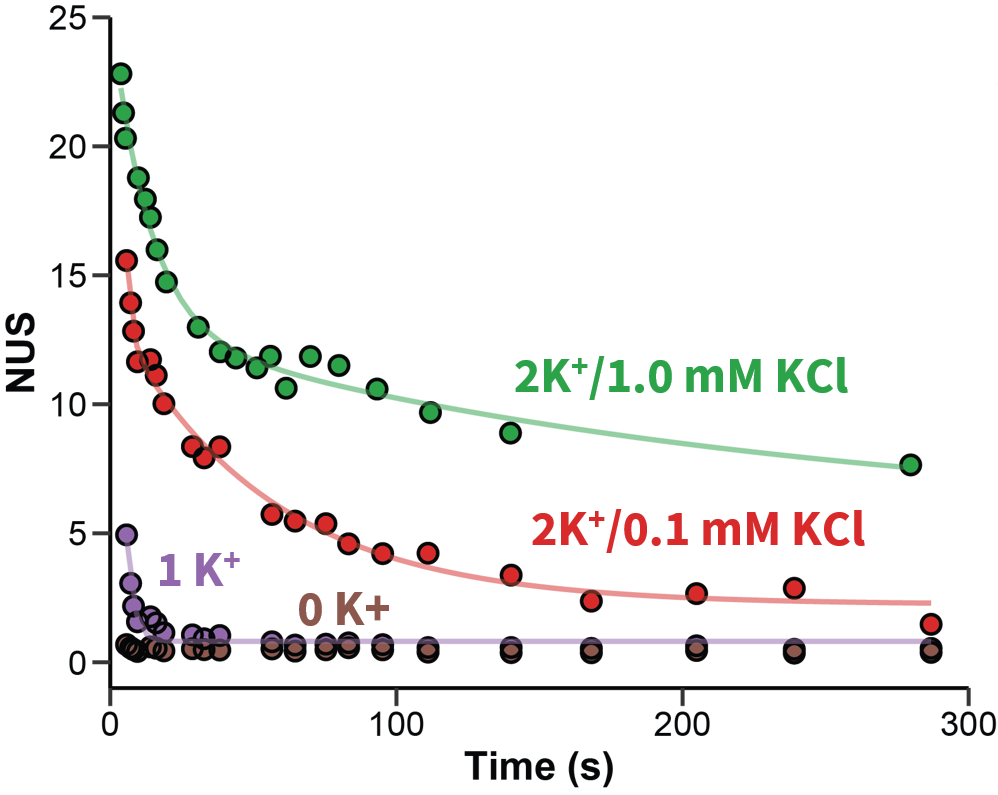
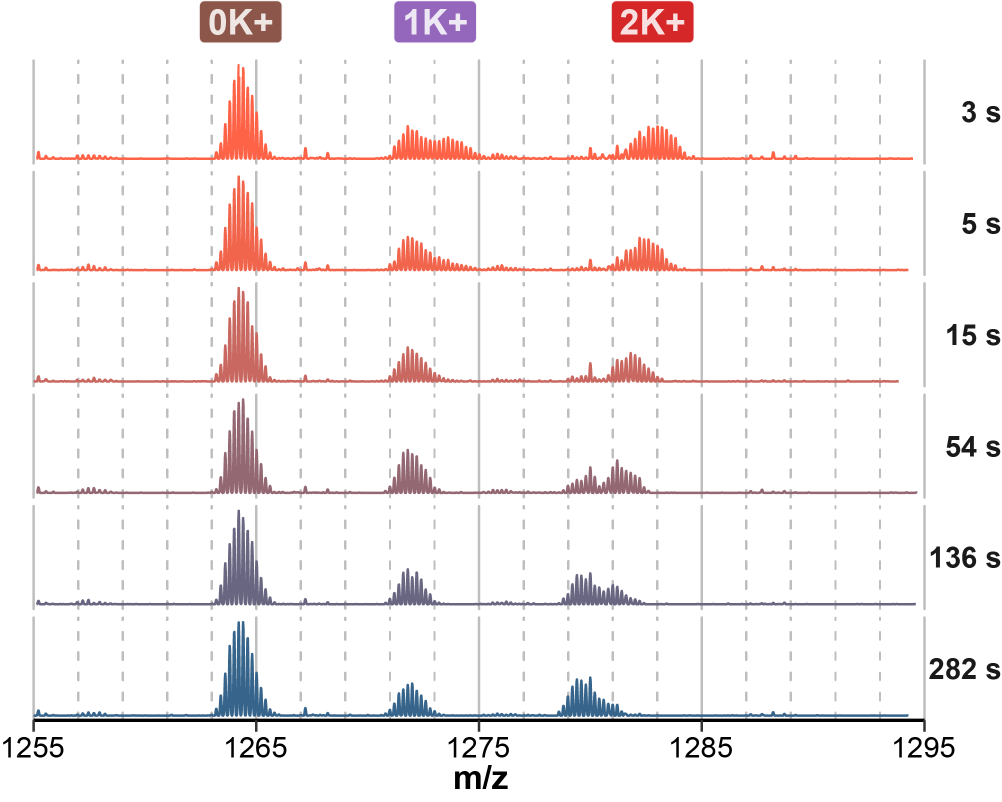
Largy and Gabelica, Anal. Chem., 2020, 92 (6), 4402–10
The number of protected sites scales with the number of tetrads

Largy, Ranz, Gabelica, J. Am. Chem. Soc. 2023, 145, 49, 26843
The number of protected sites scales with the number of tetrads

Largy, Ranz, Gabelica, J. Am. Chem. Soc. 2023, 145, 49, 26843
The number of protected sites scales with the number of tetrads


Largy, Ranz, Gabelica, J. Am. Chem. Soc. 2023, 145, 49, 26843
Exchange protection scales with stability
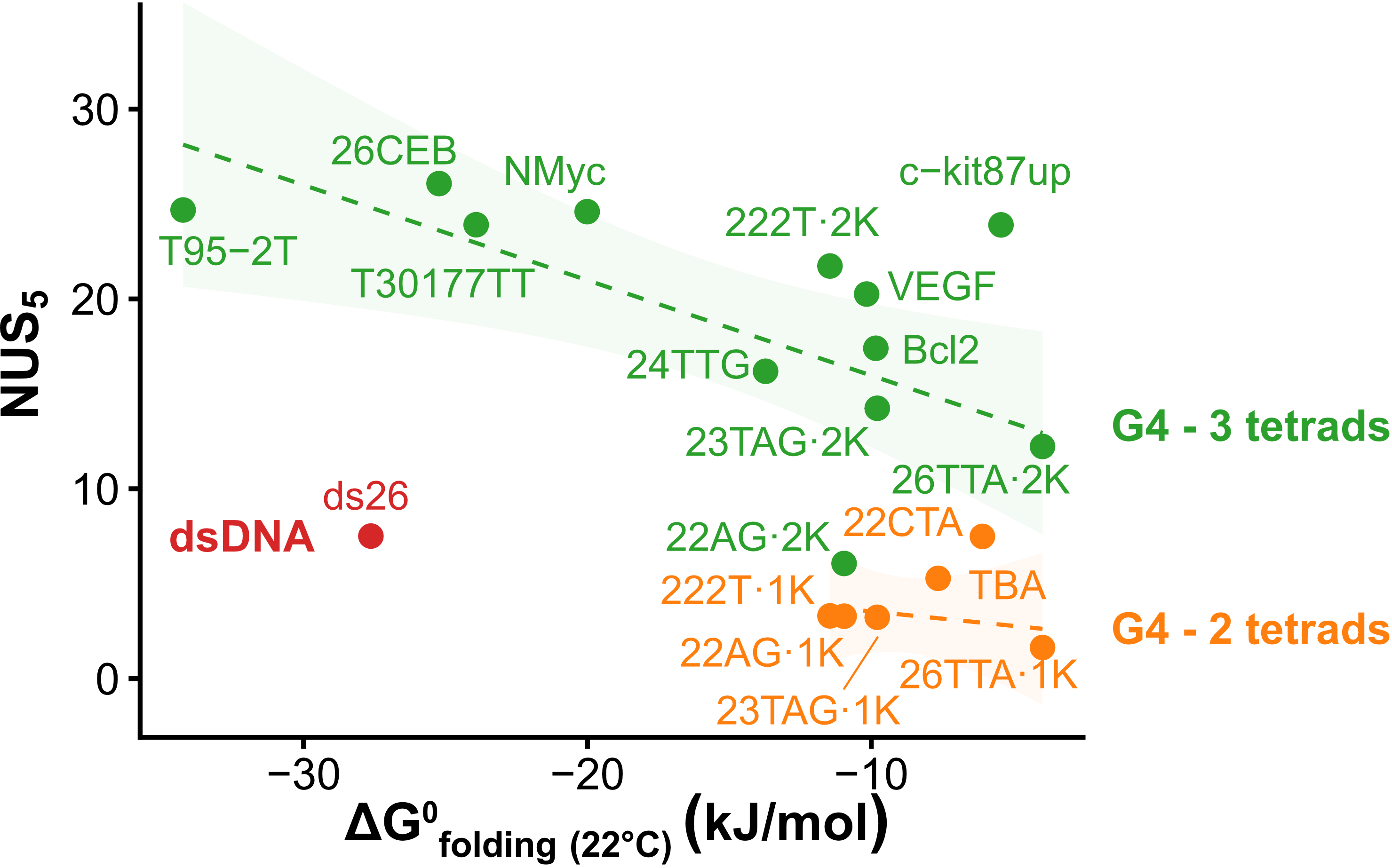
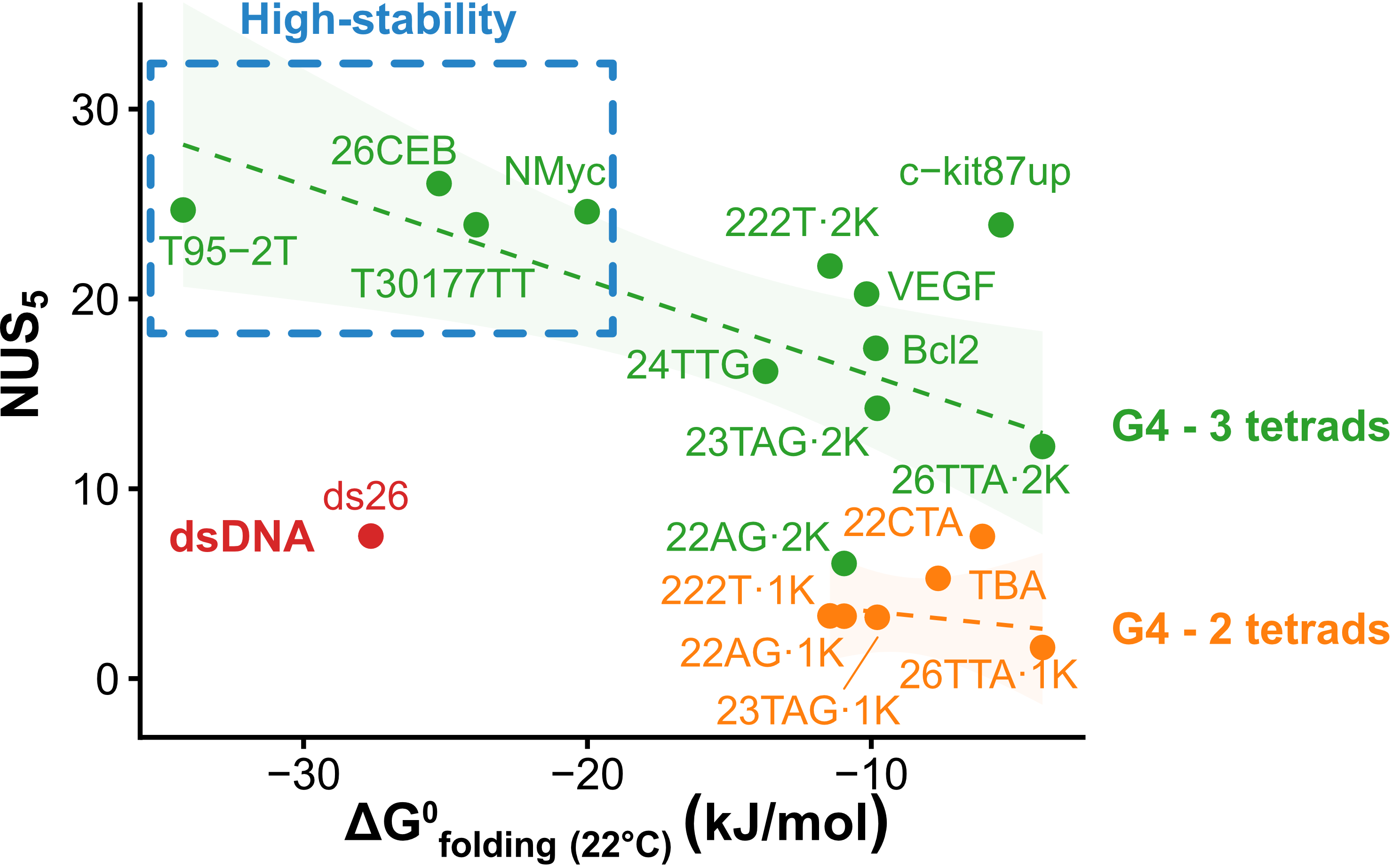
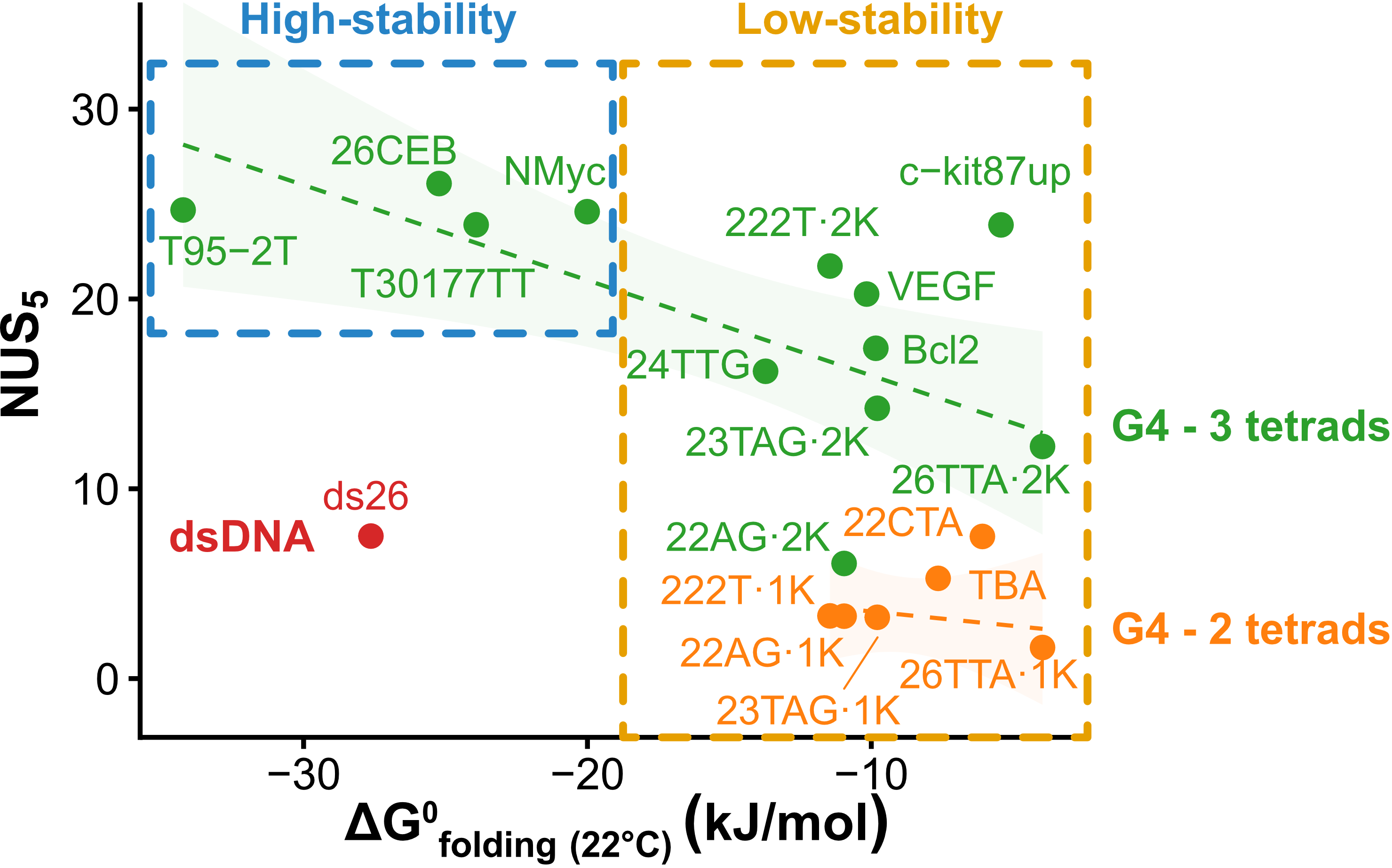
Largy, Ranz, Gabelica, J. Am. Chem. Soc. 2023, 145, 49, 26843
Higher stability G4s exchange via local fluctuations
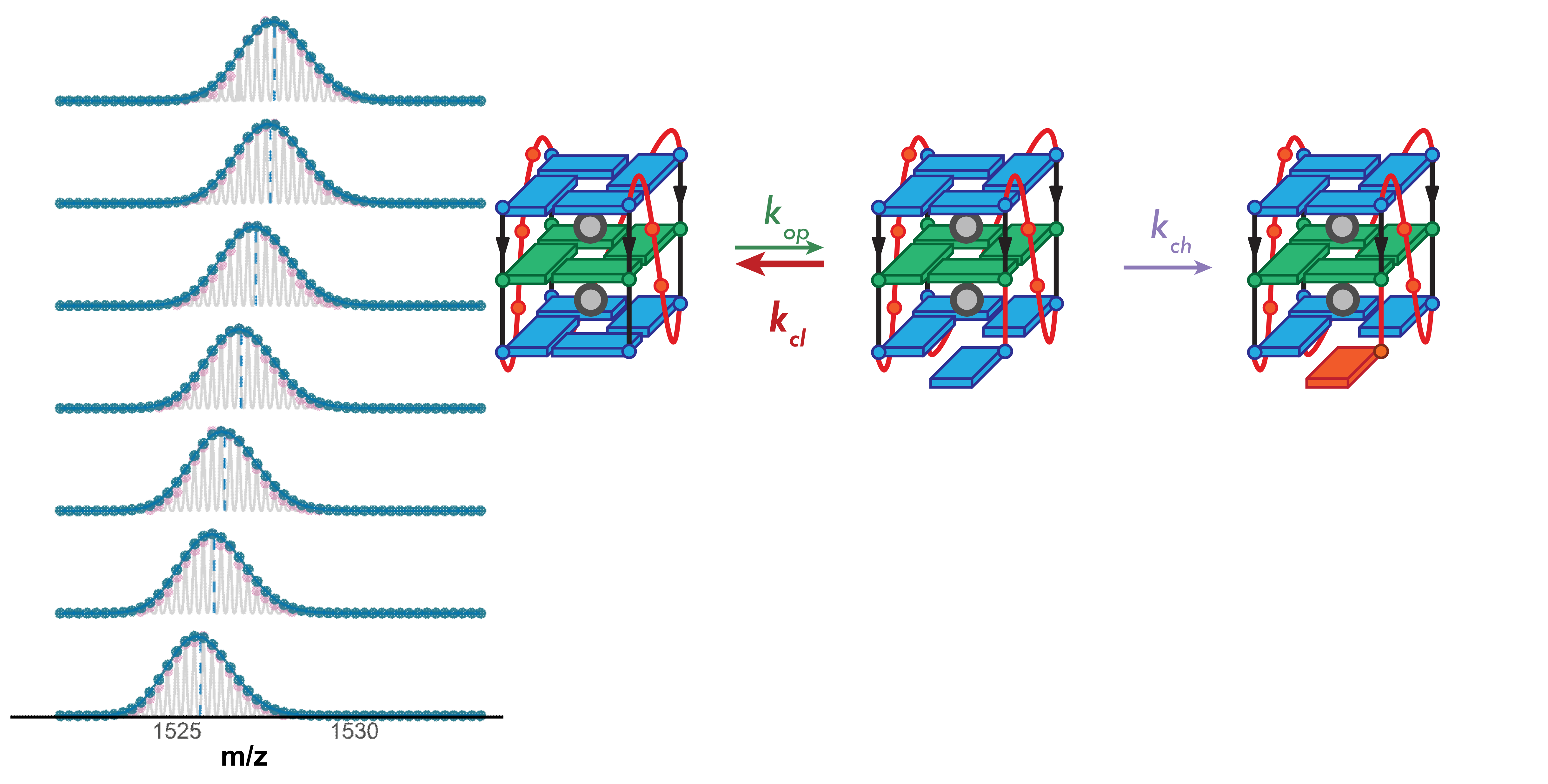
- Short-lived local fluctuations
- Local exchange: “EX2”
- Monomodal isotopic distributions
Largy, Ranz, Gabelica, J. Am. Chem. Soc. 2023, 145, 49, 26843
Lower stability G4s exchange via an unfolded conformer
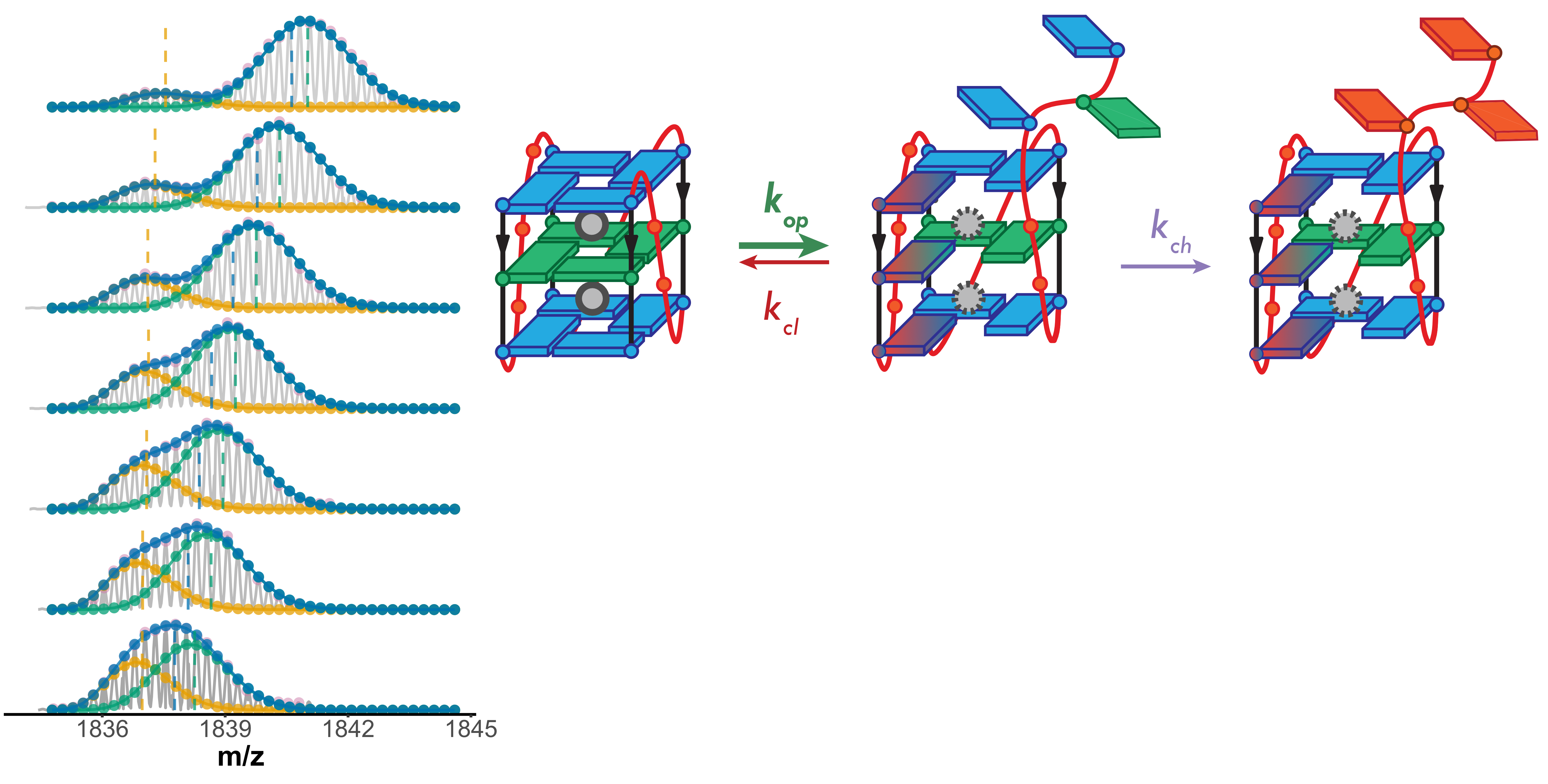
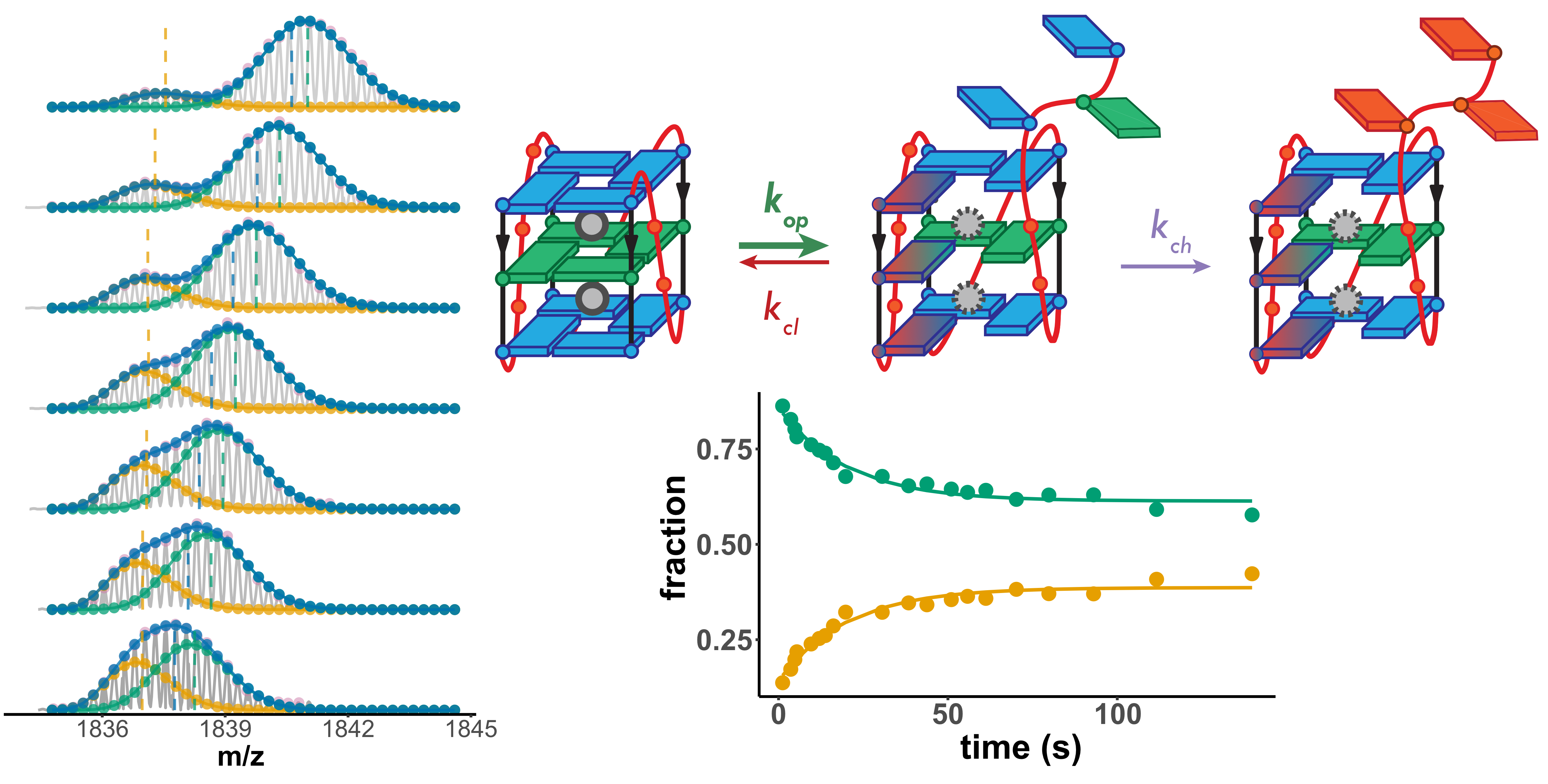
- Longer-lived unfolded conformers
- Global exchange: “EX1”
- Multimodal isotopic distributions
- Each molecule visiting the exchange-competent conformation is revealed
Largy, Ranz, Gabelica, J. Am. Chem. Soc. 2023, 145, 49, 26843
Exchange-competent species resemble folding intermediates


Largy, Ranz, Gabelica, J. Am. Chem. Soc. 2023, 145, 49, 26843
Drift tube IMS
- Collisions between ions and drift gas
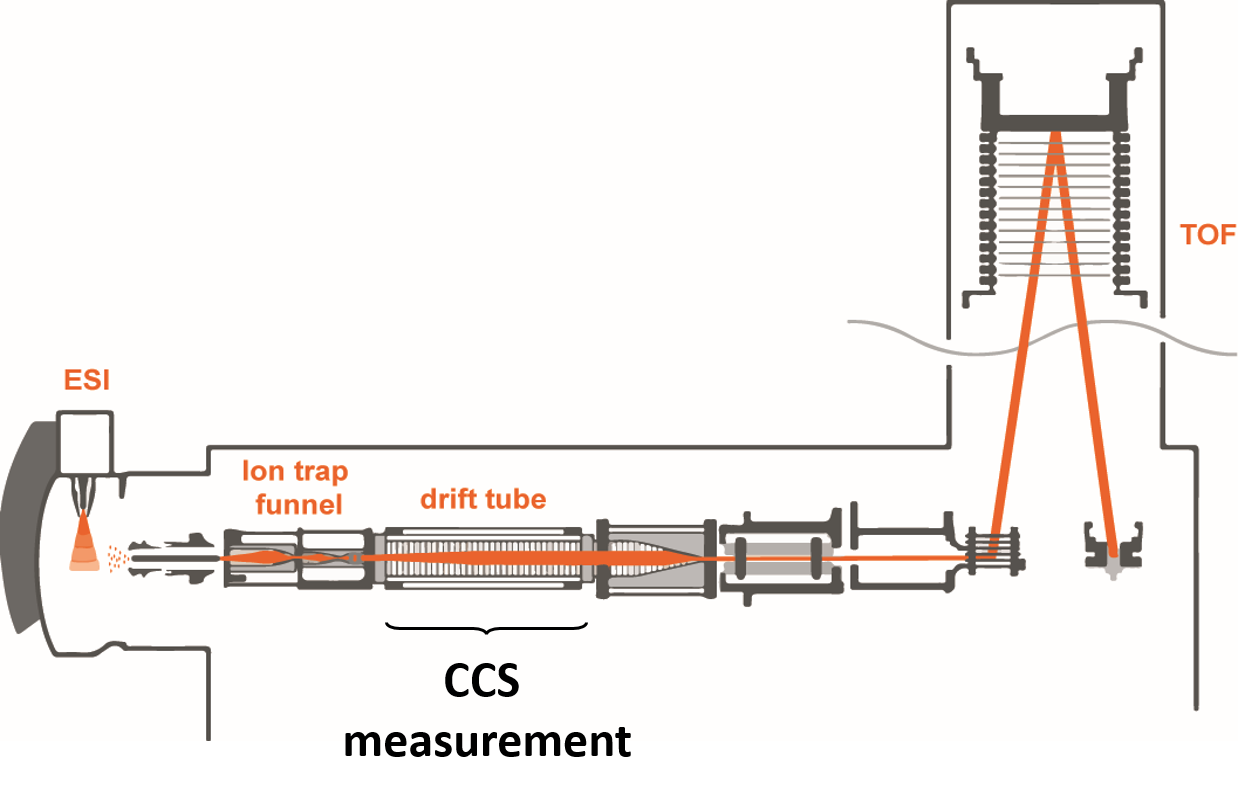
Drift tube IMS
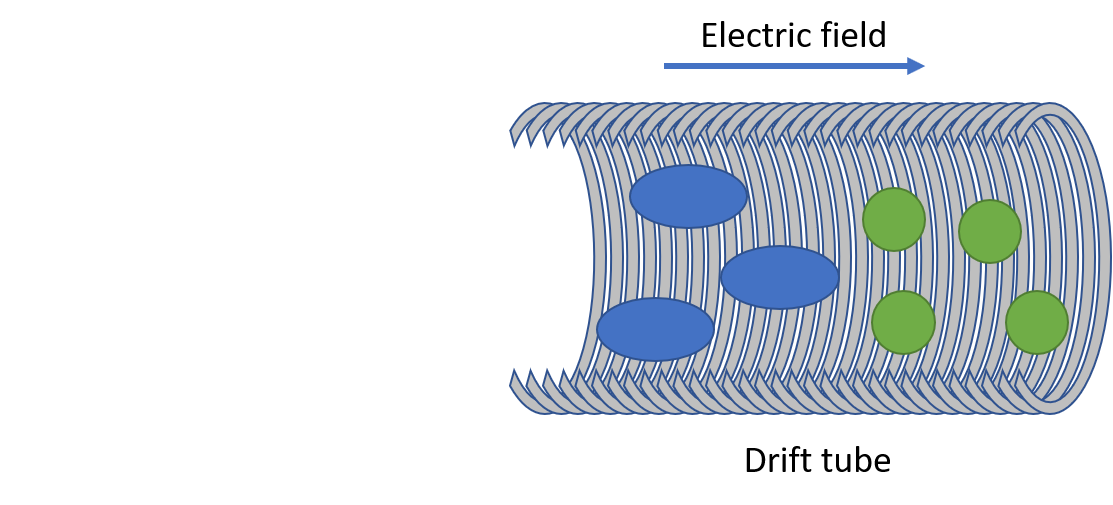
- Collisions between ions and drift gas
- Compact structures travel faster
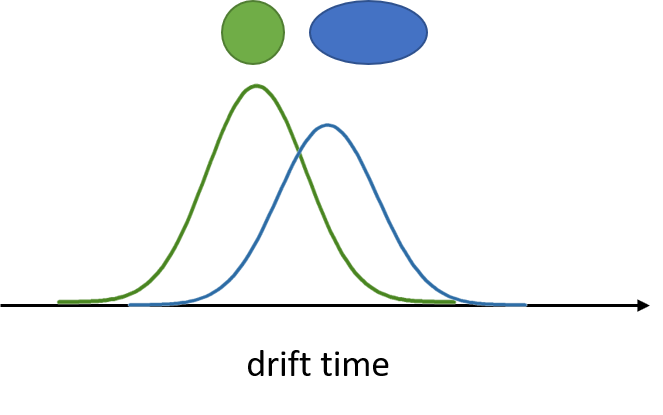
Separation of conformers of same mass
Minor unfolding conformers can be revealed by HDX/IMS

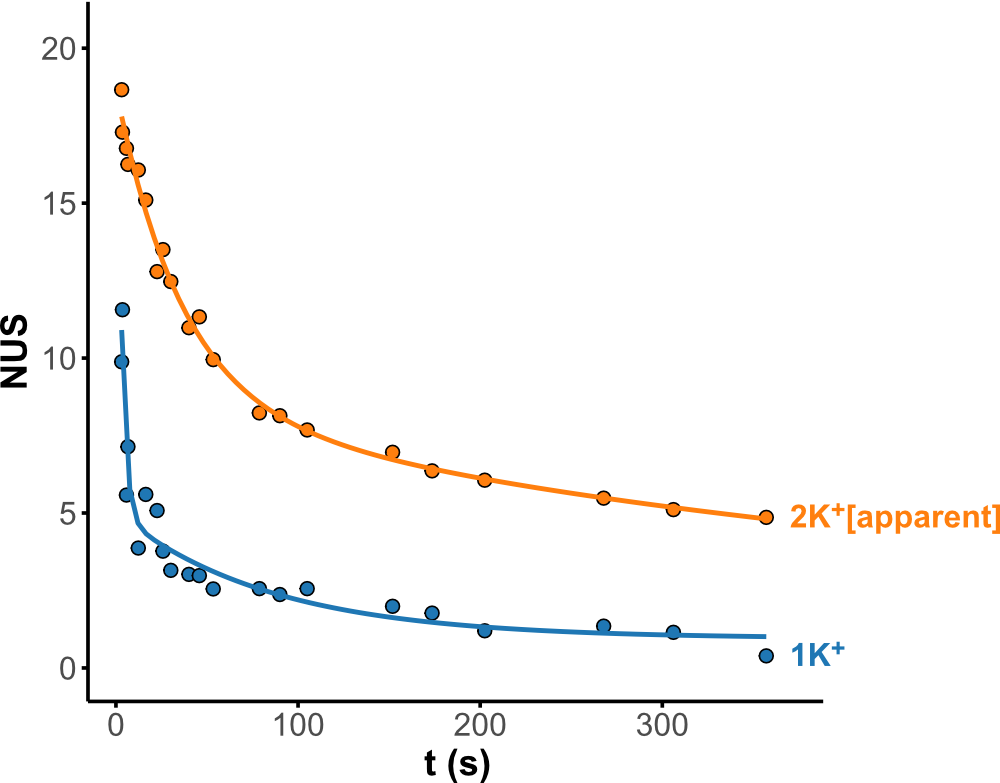

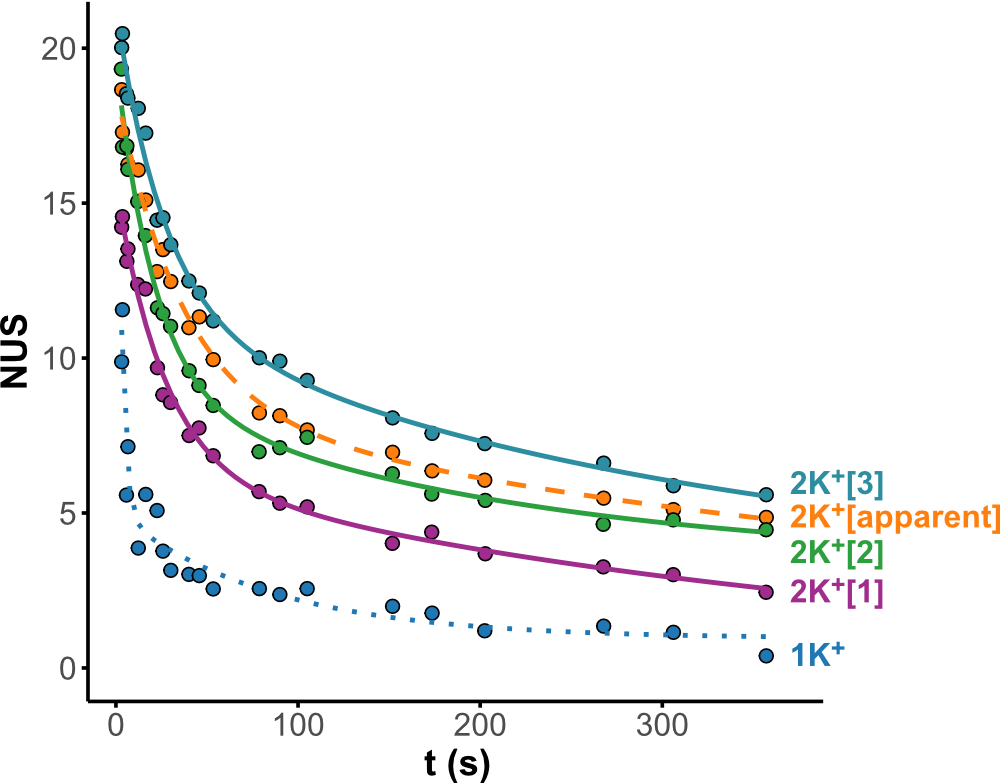
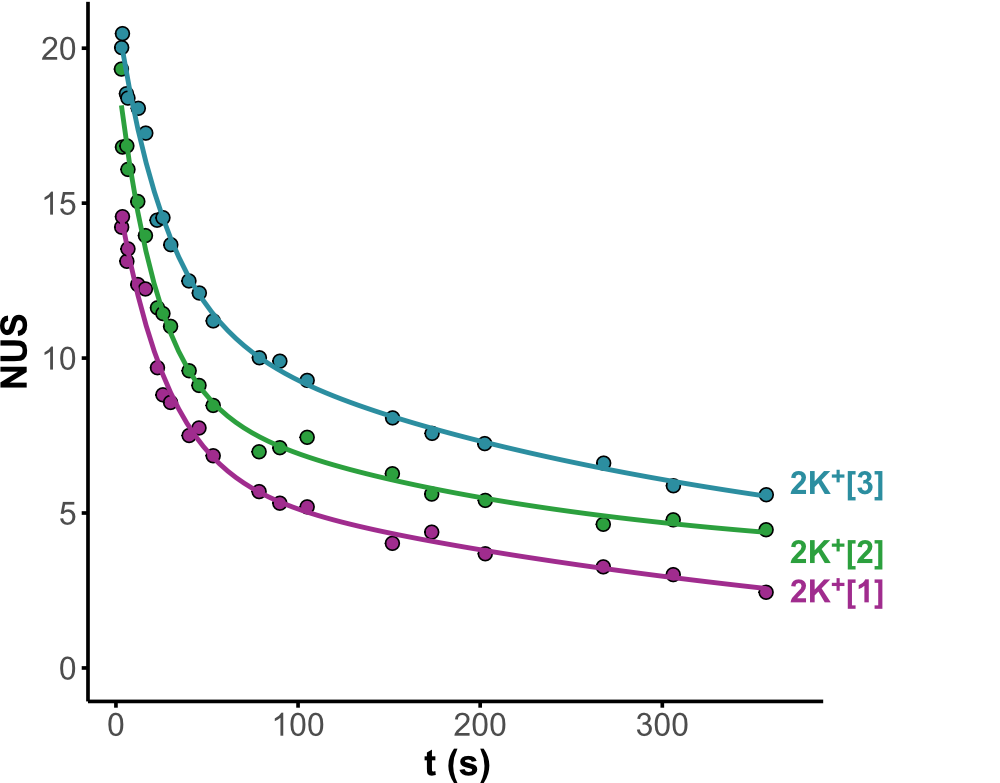

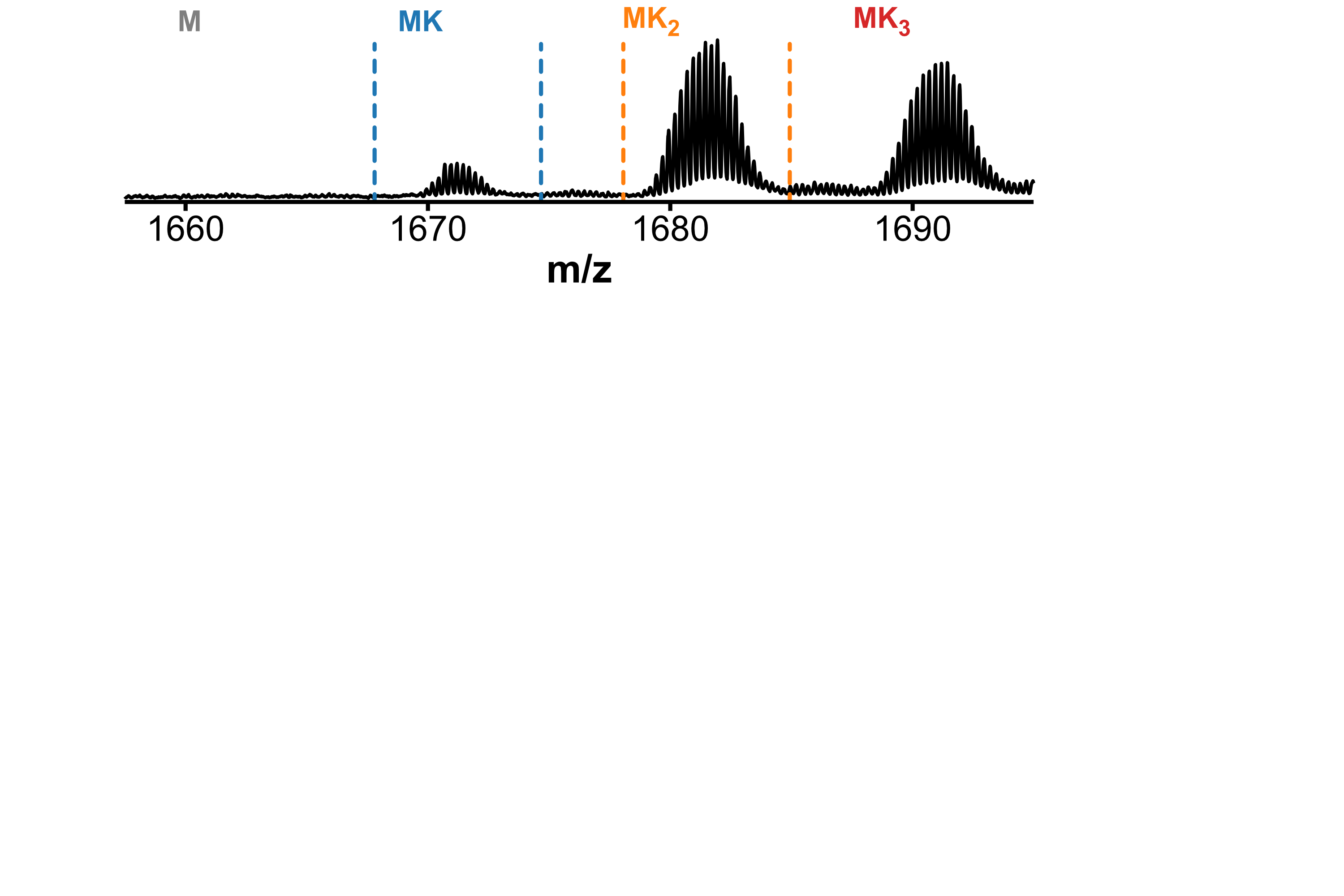
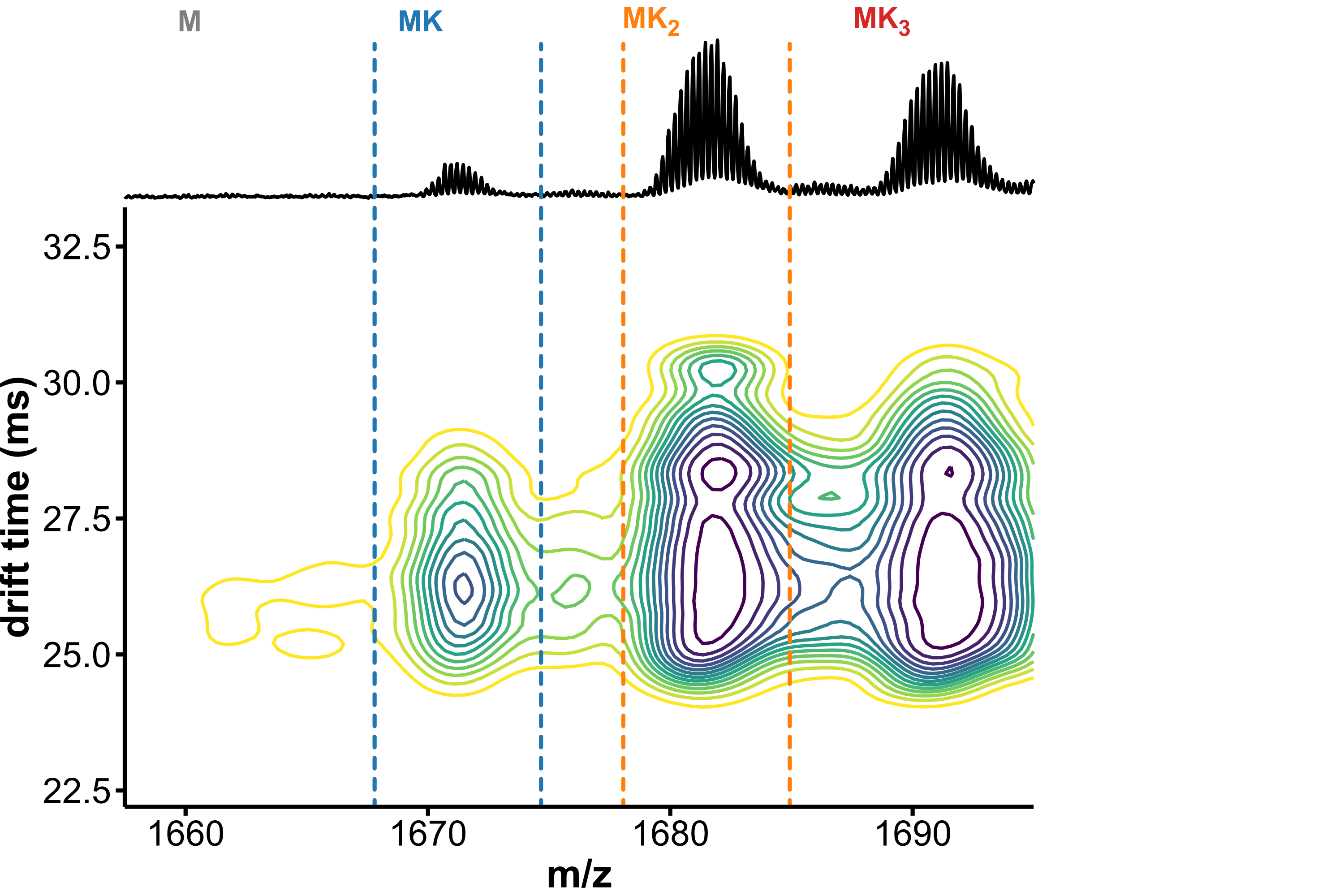
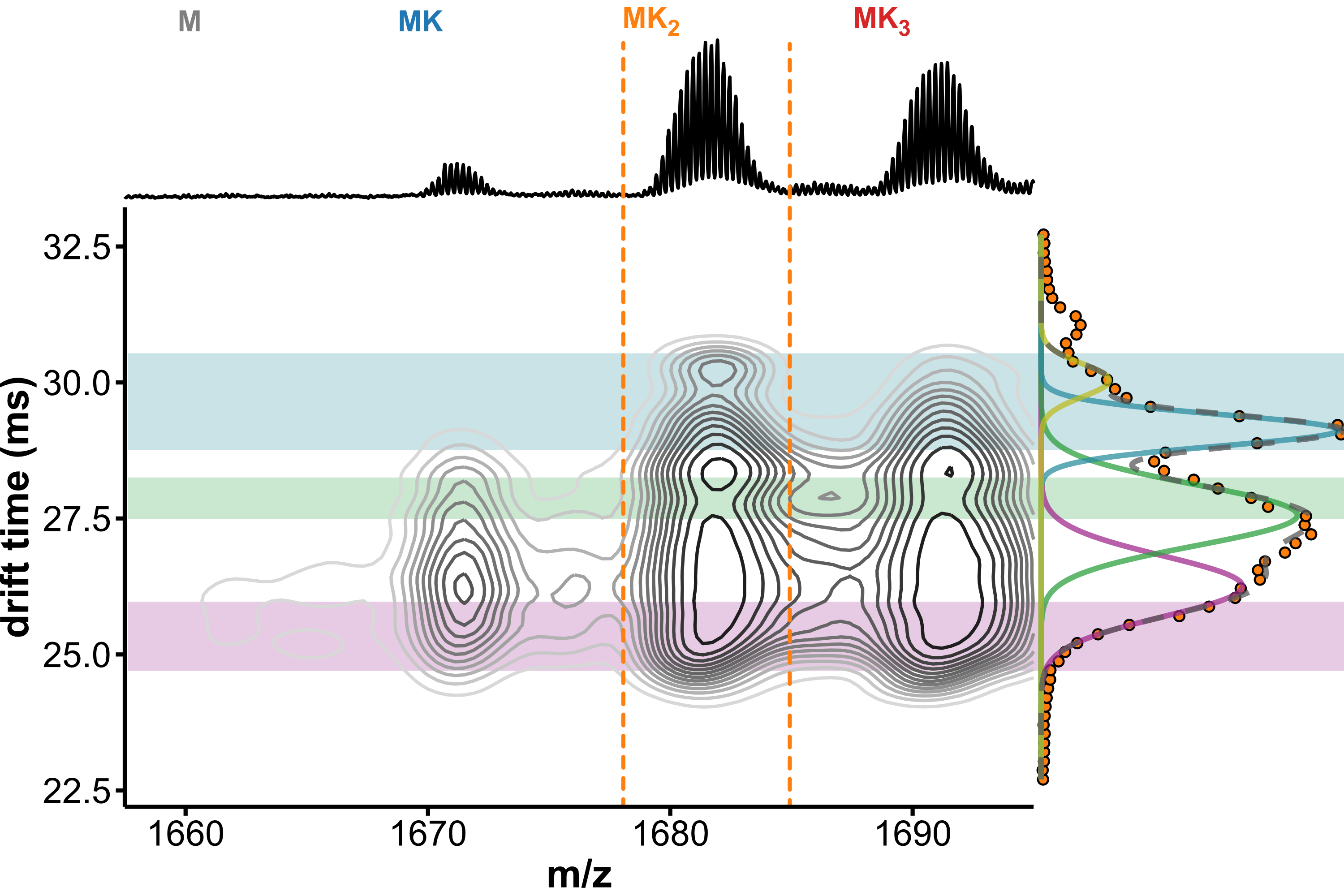

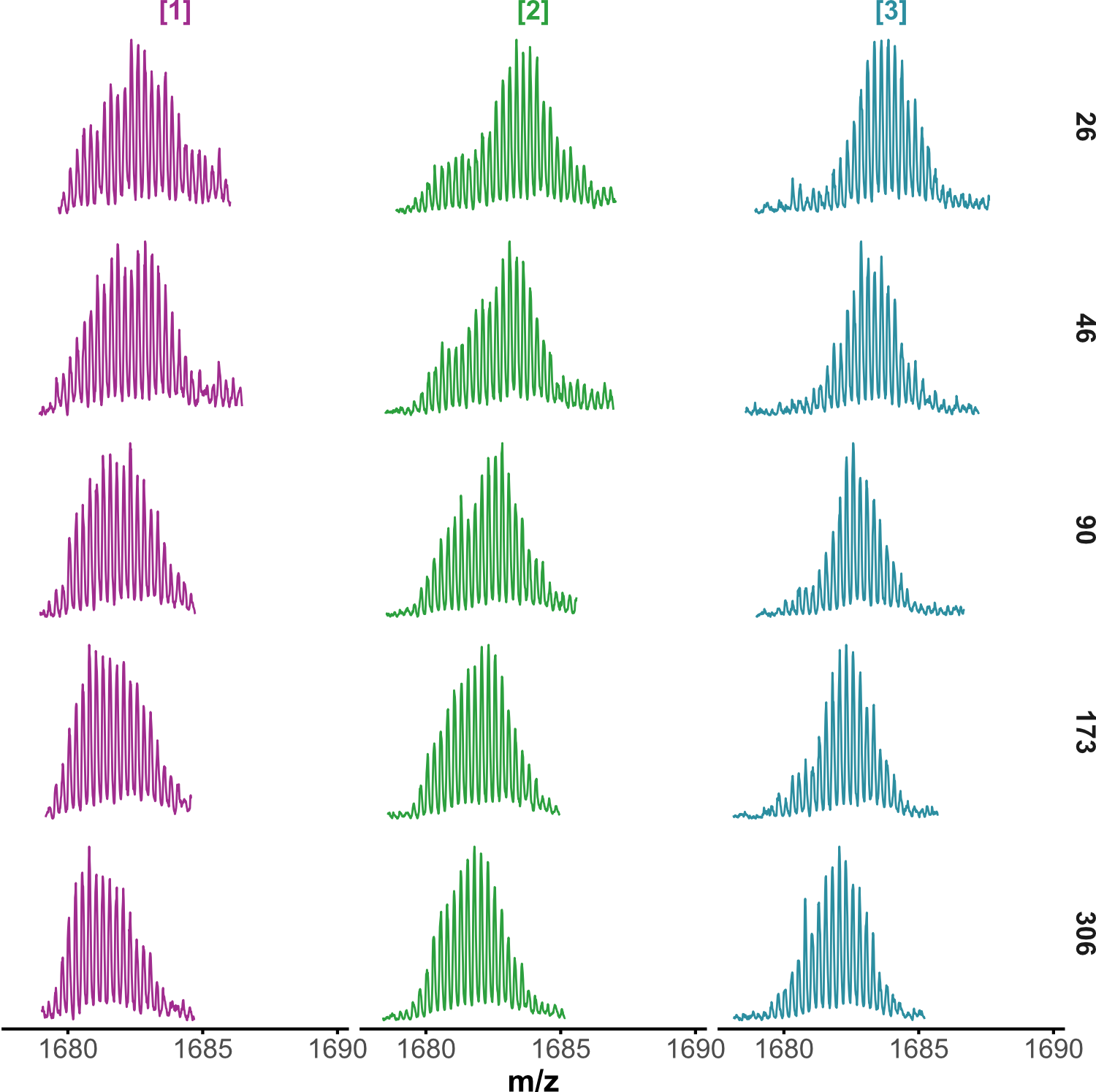
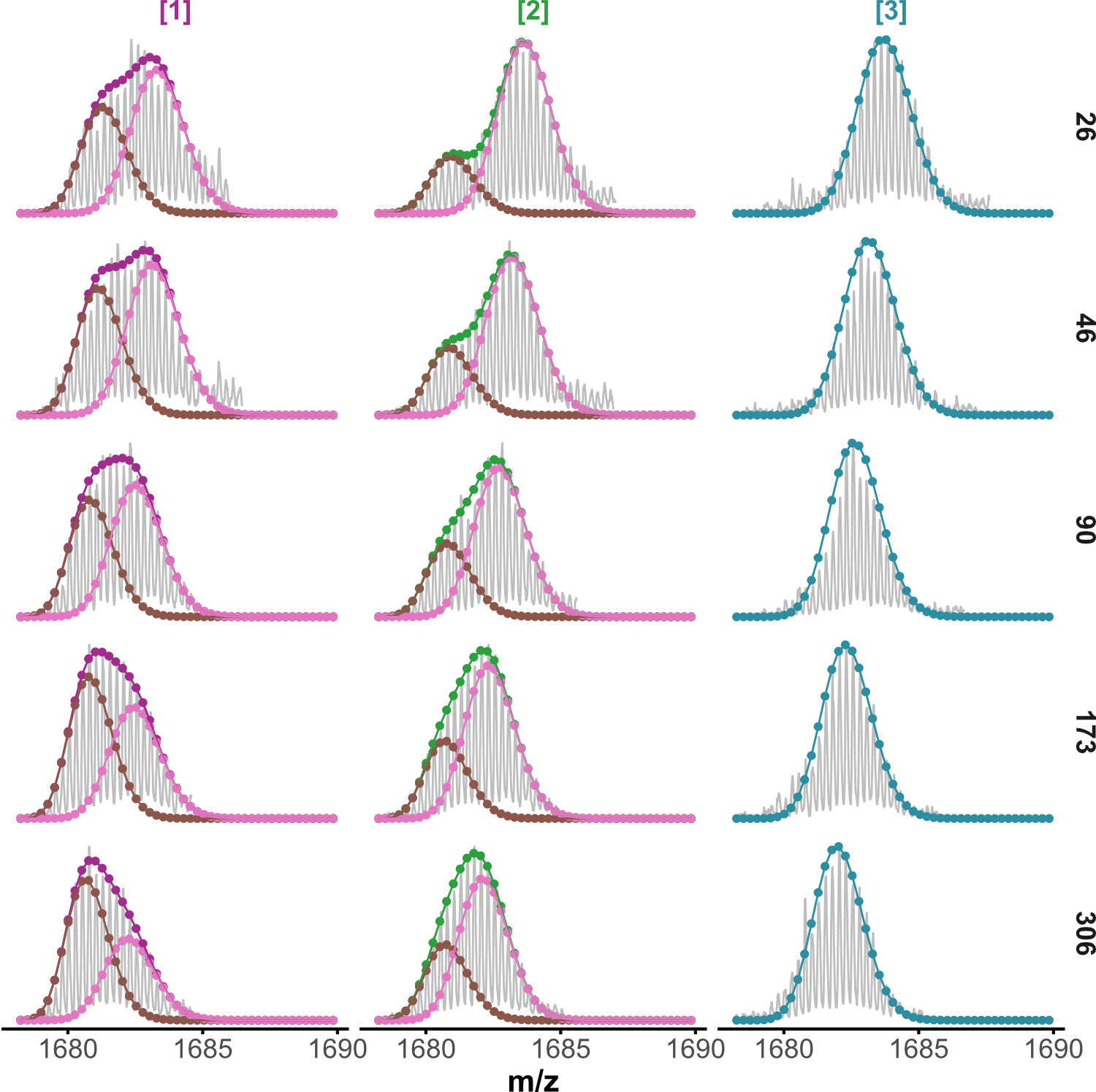



HDX kinetics inform on binding modes
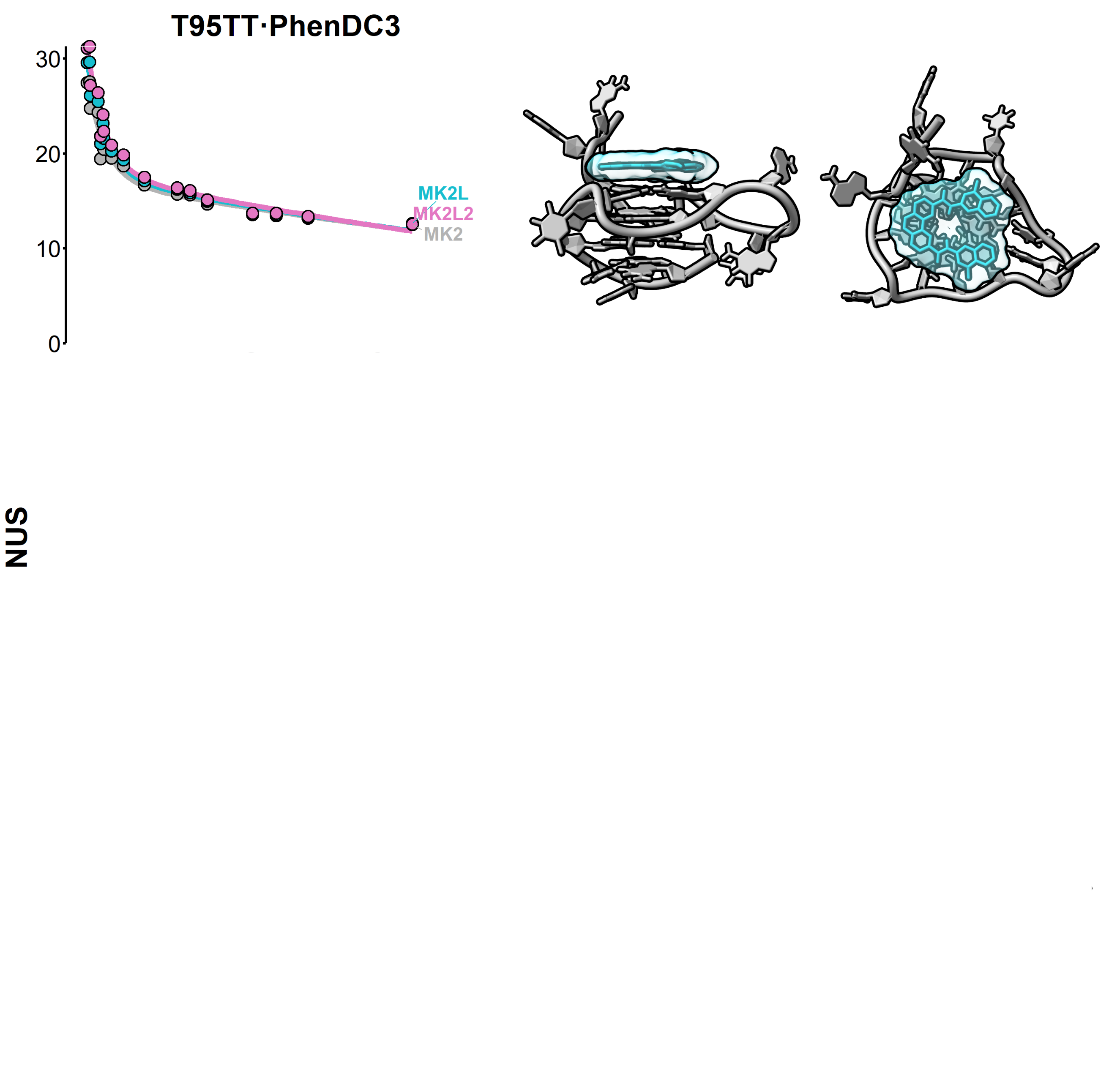
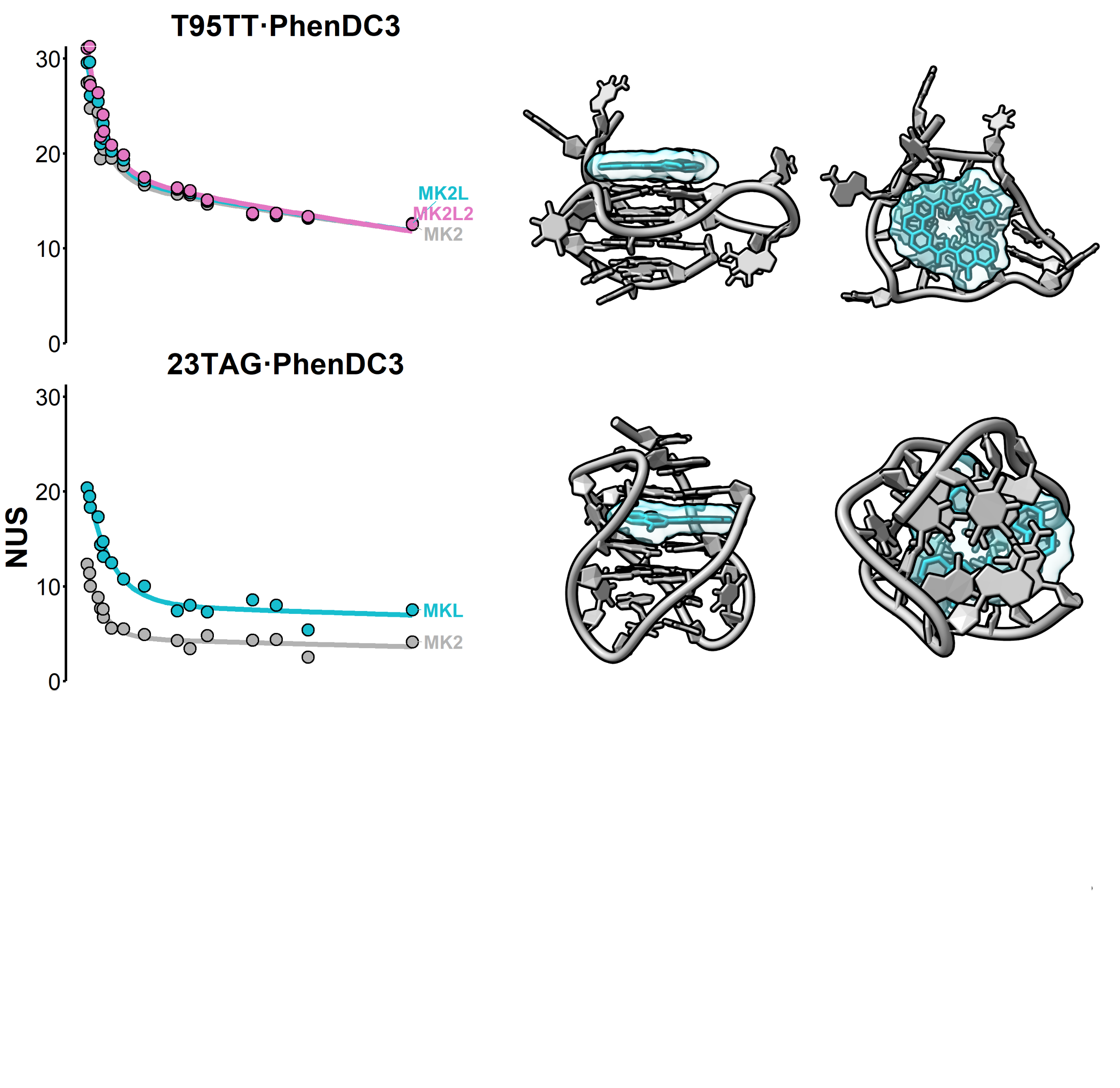
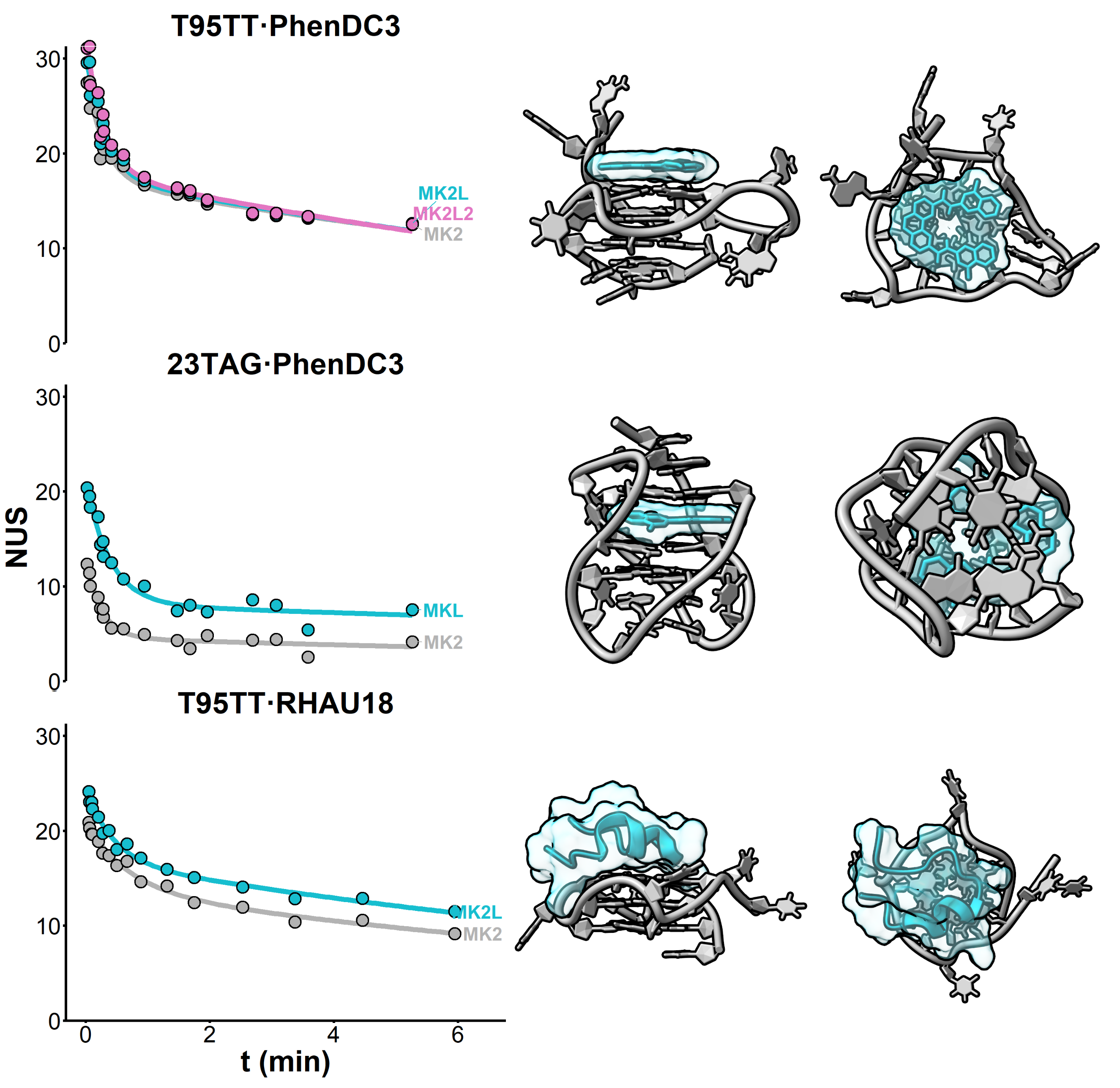


Computational approaches
Eps2Fold: structural insights from UV spectra
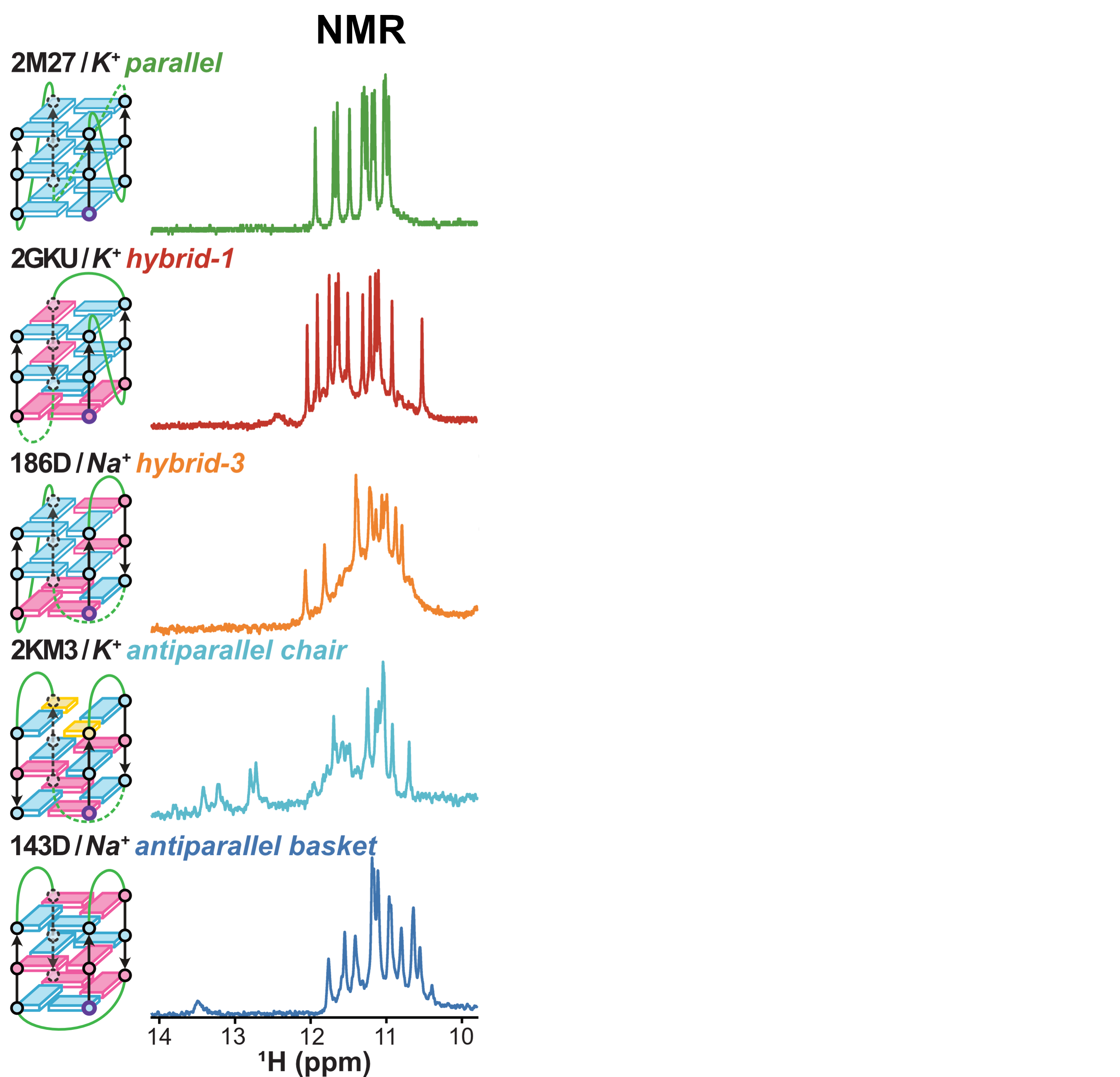
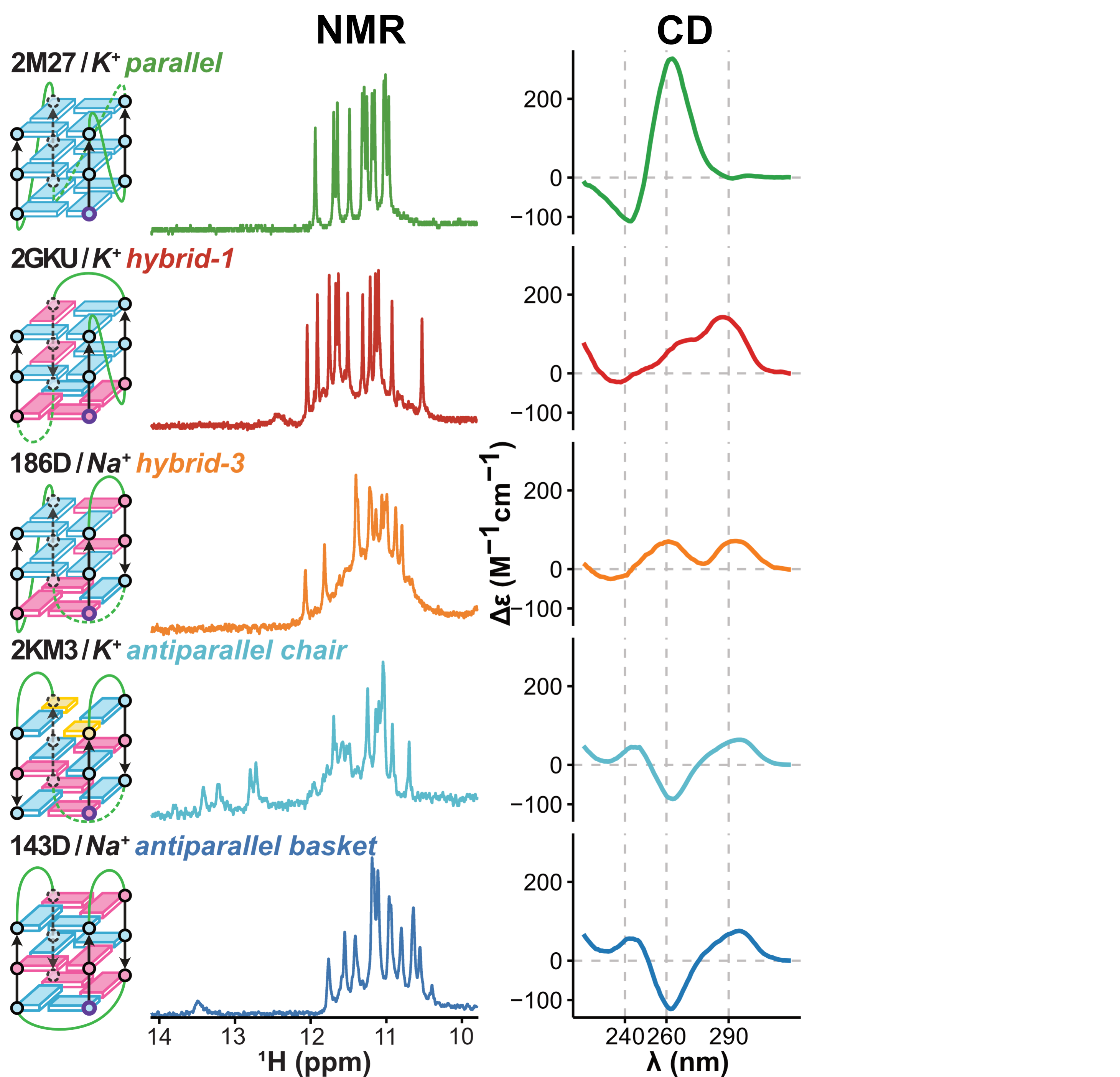
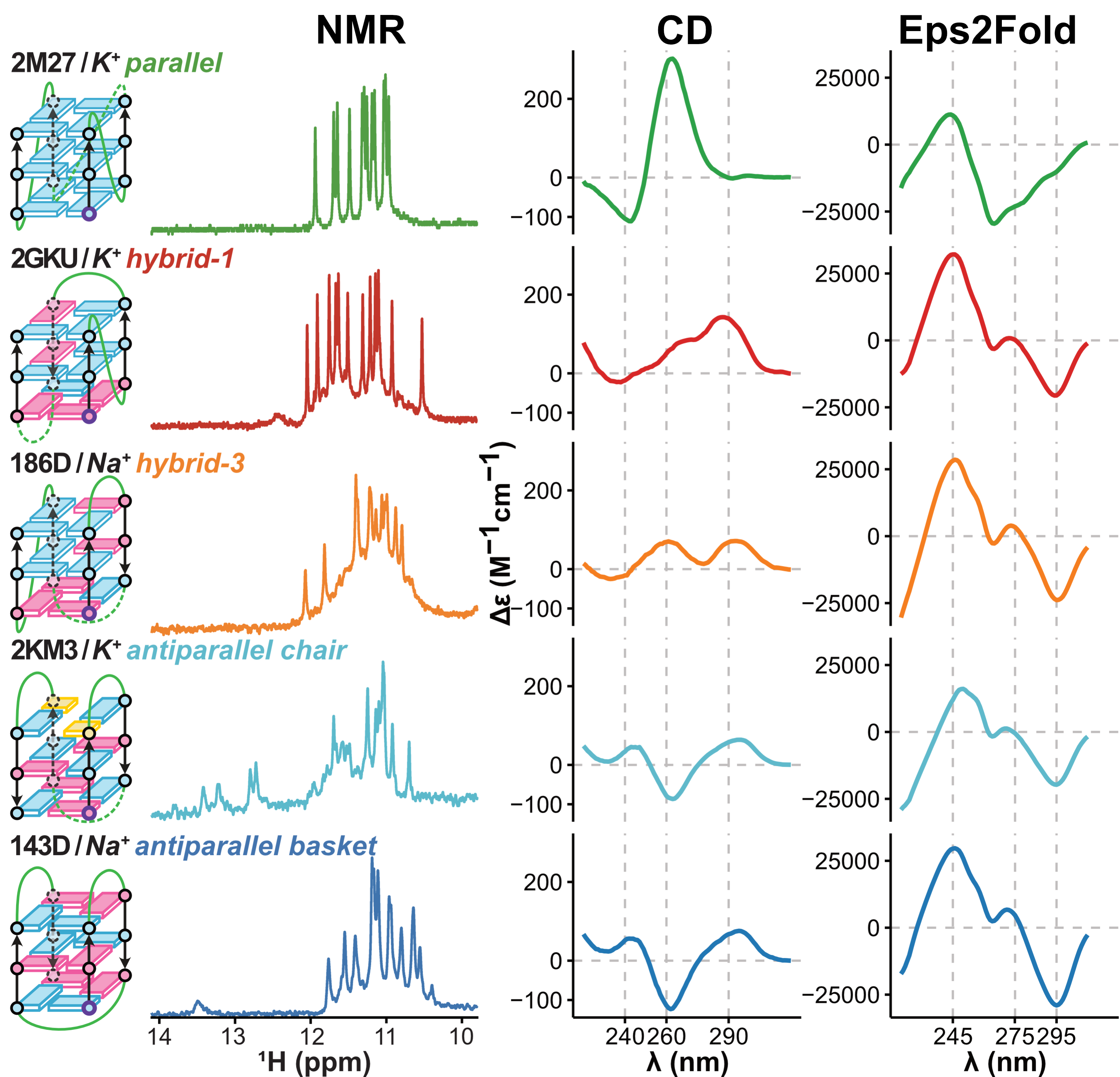

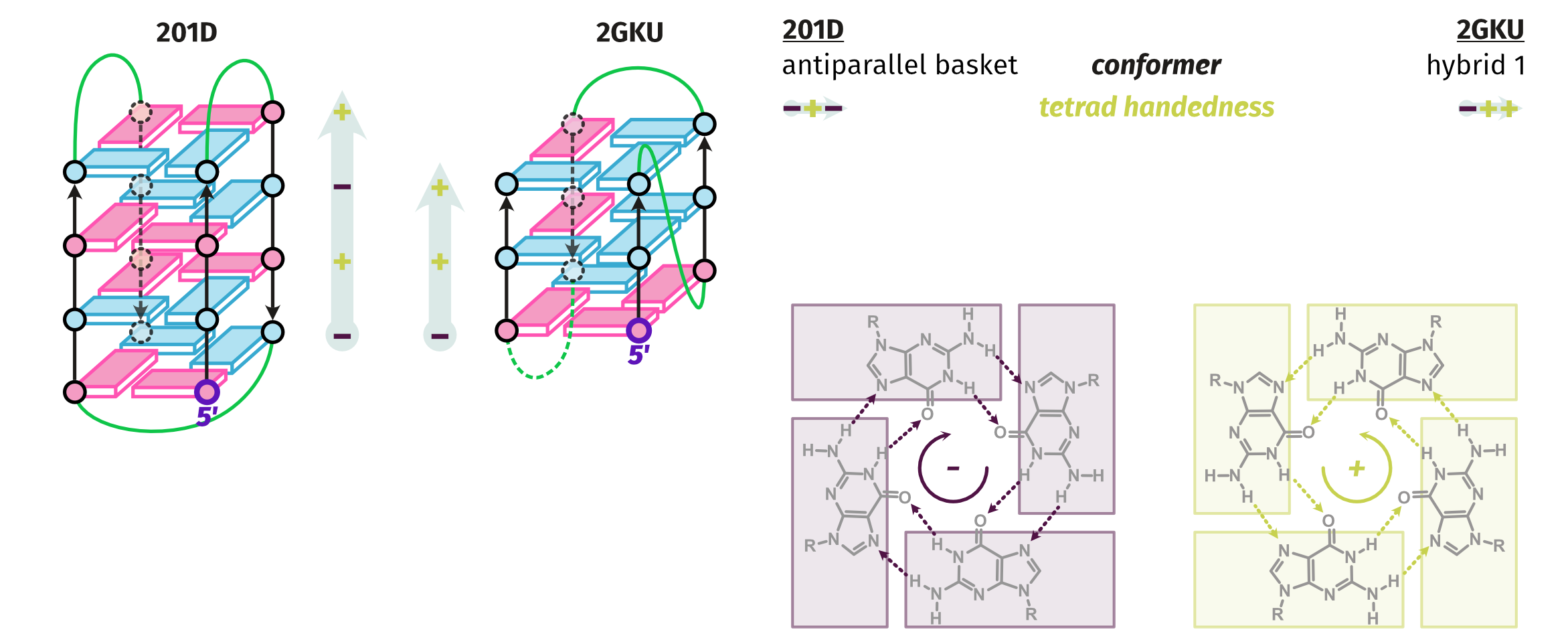
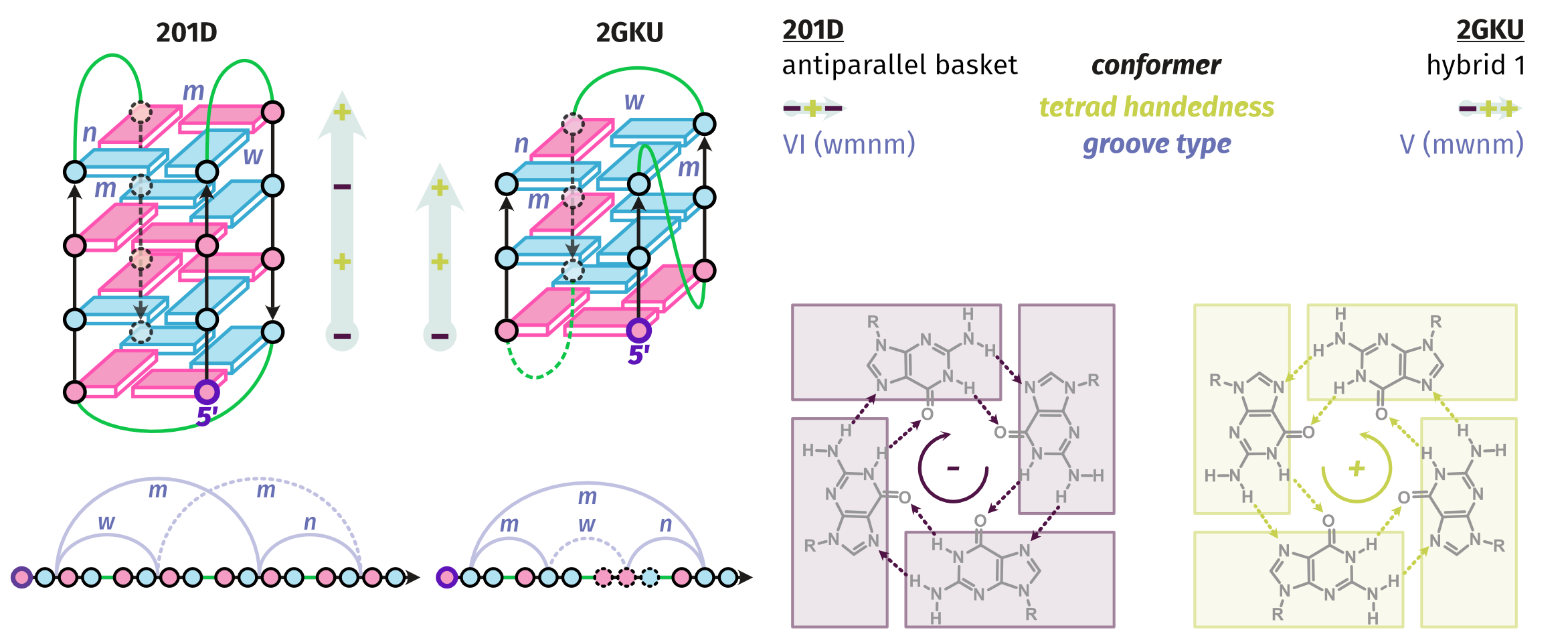
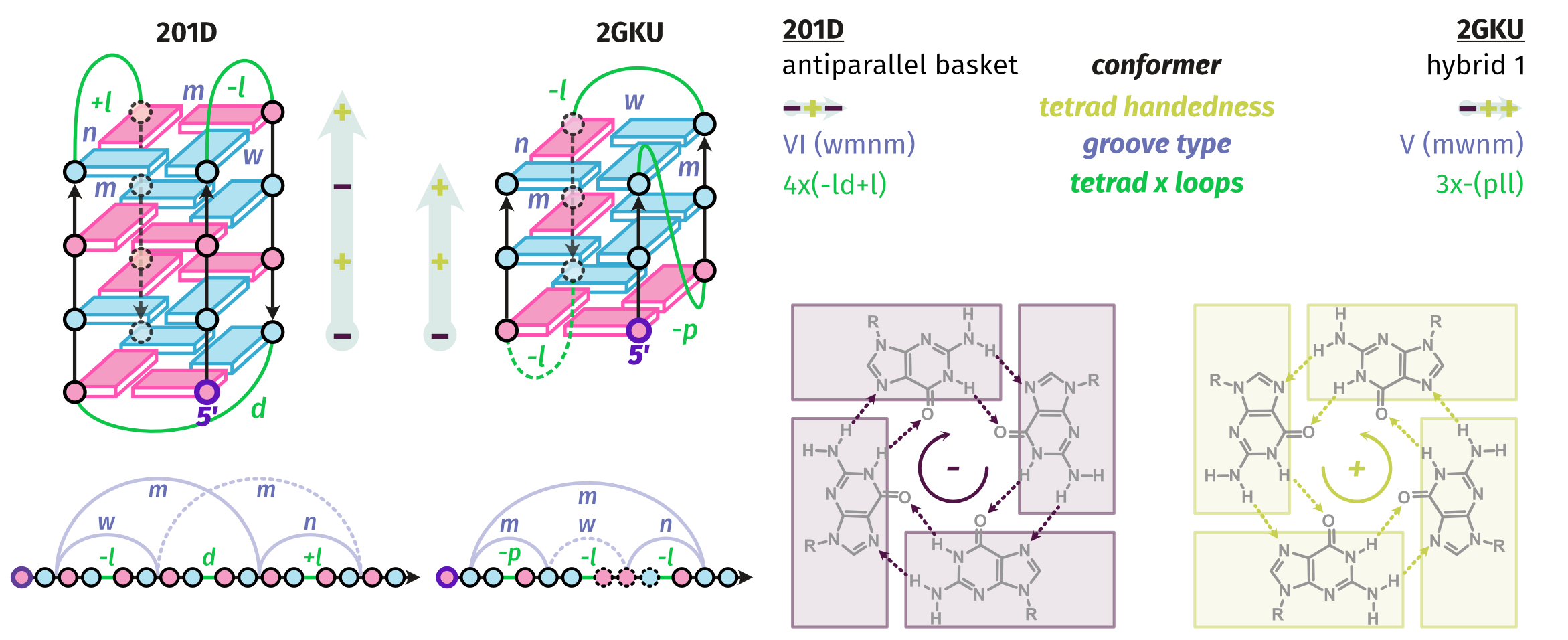
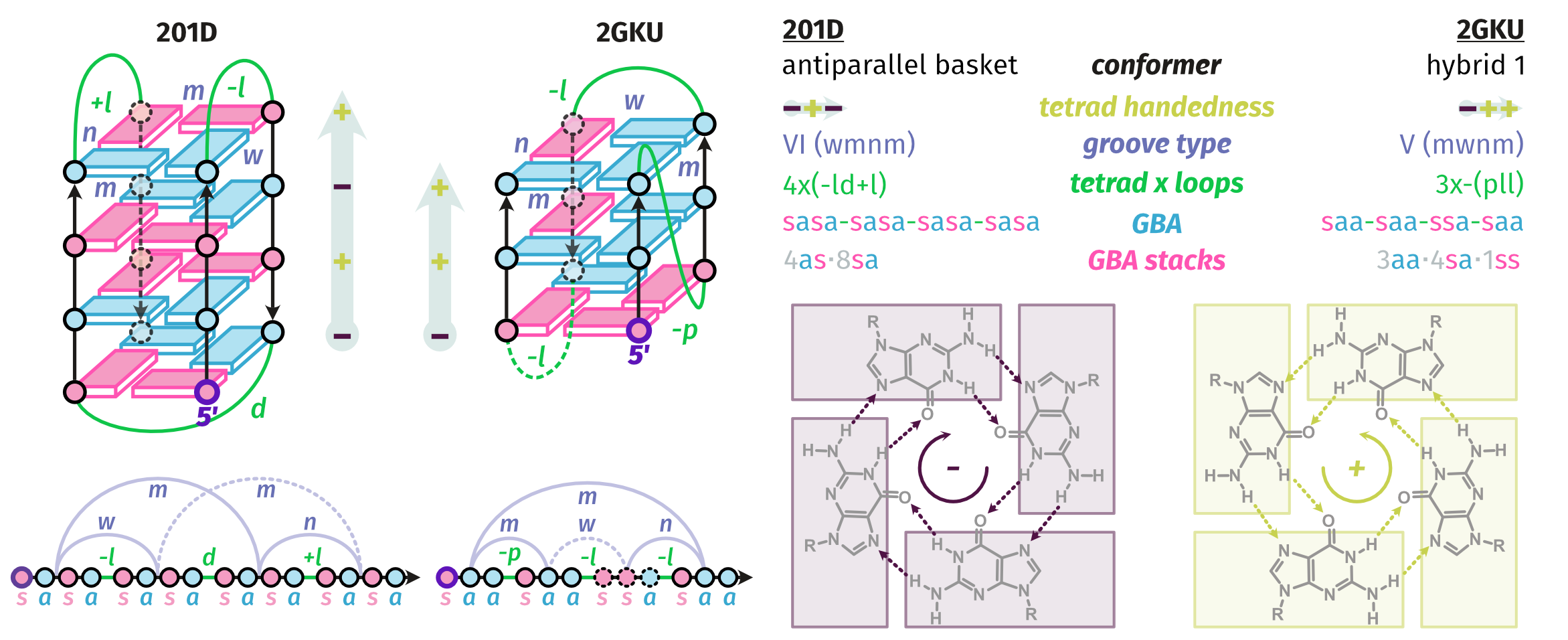
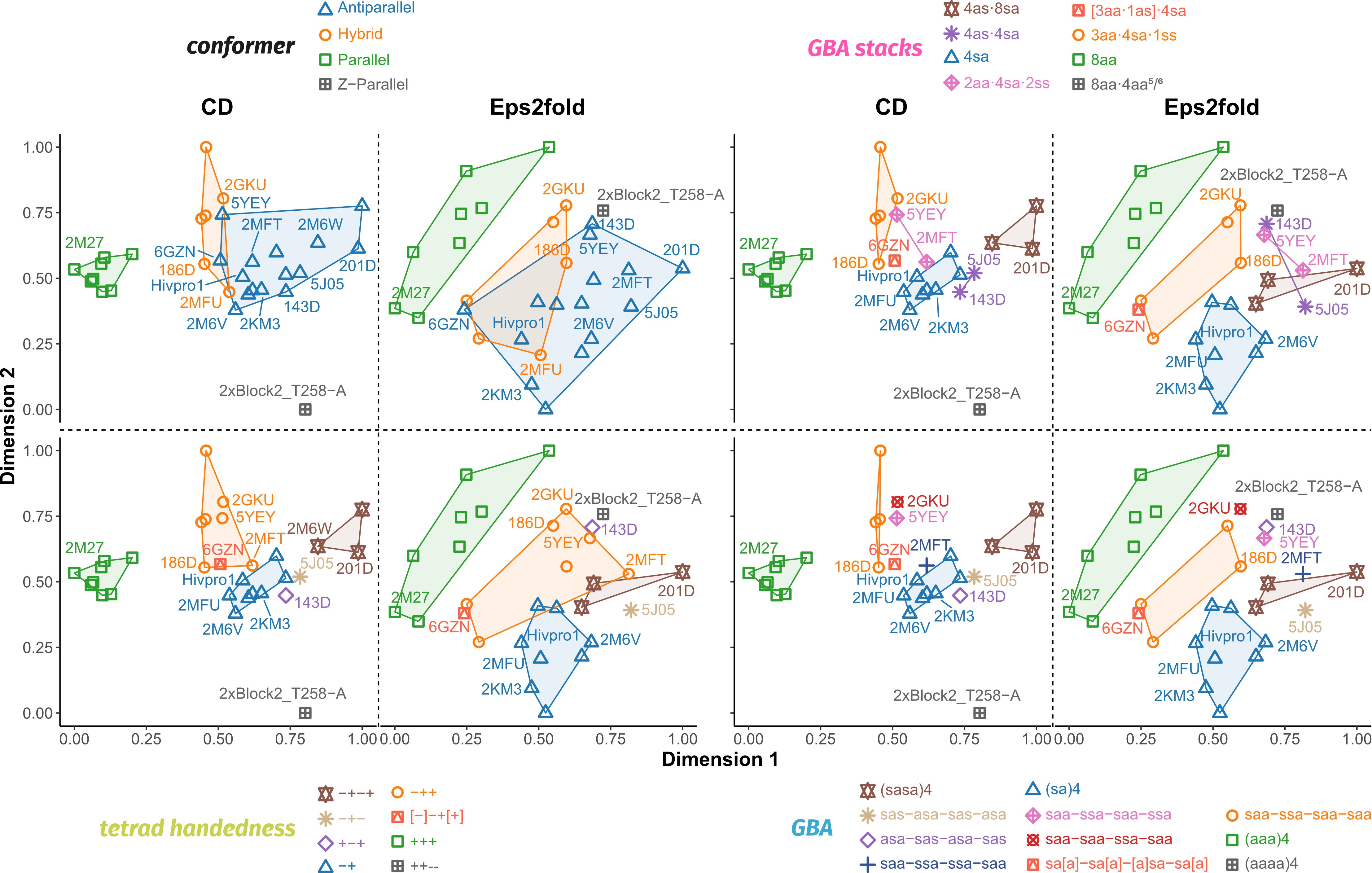
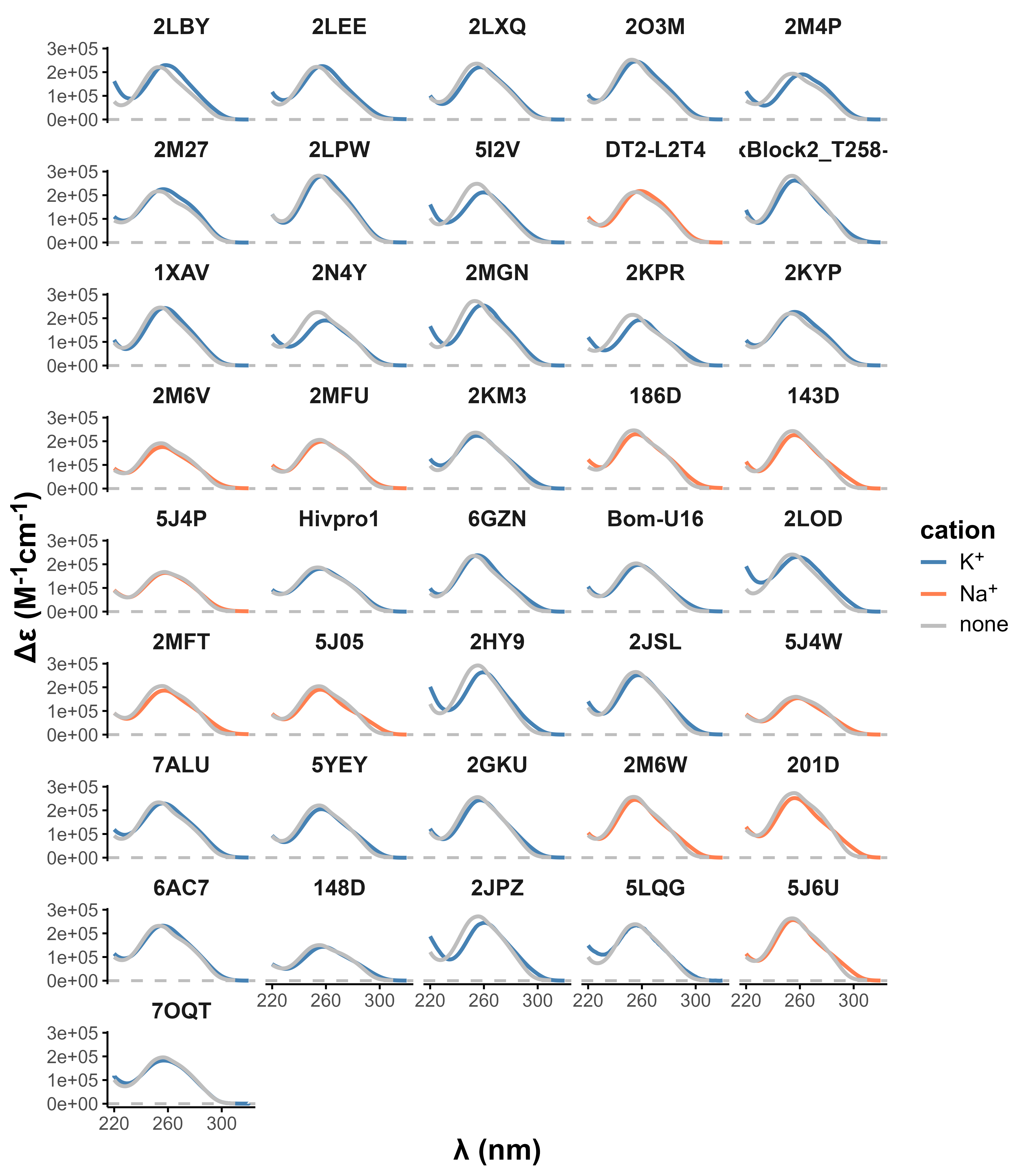
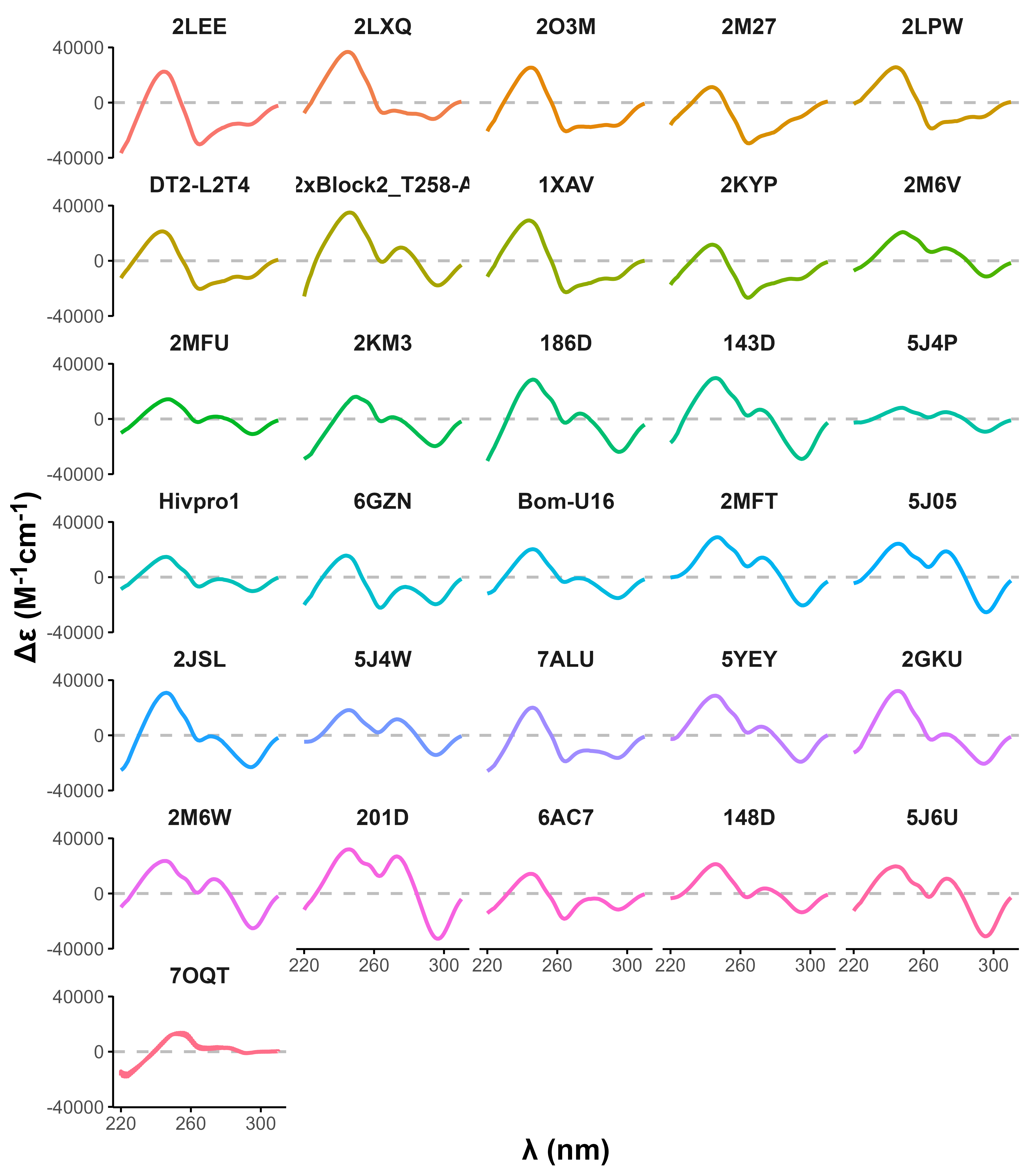


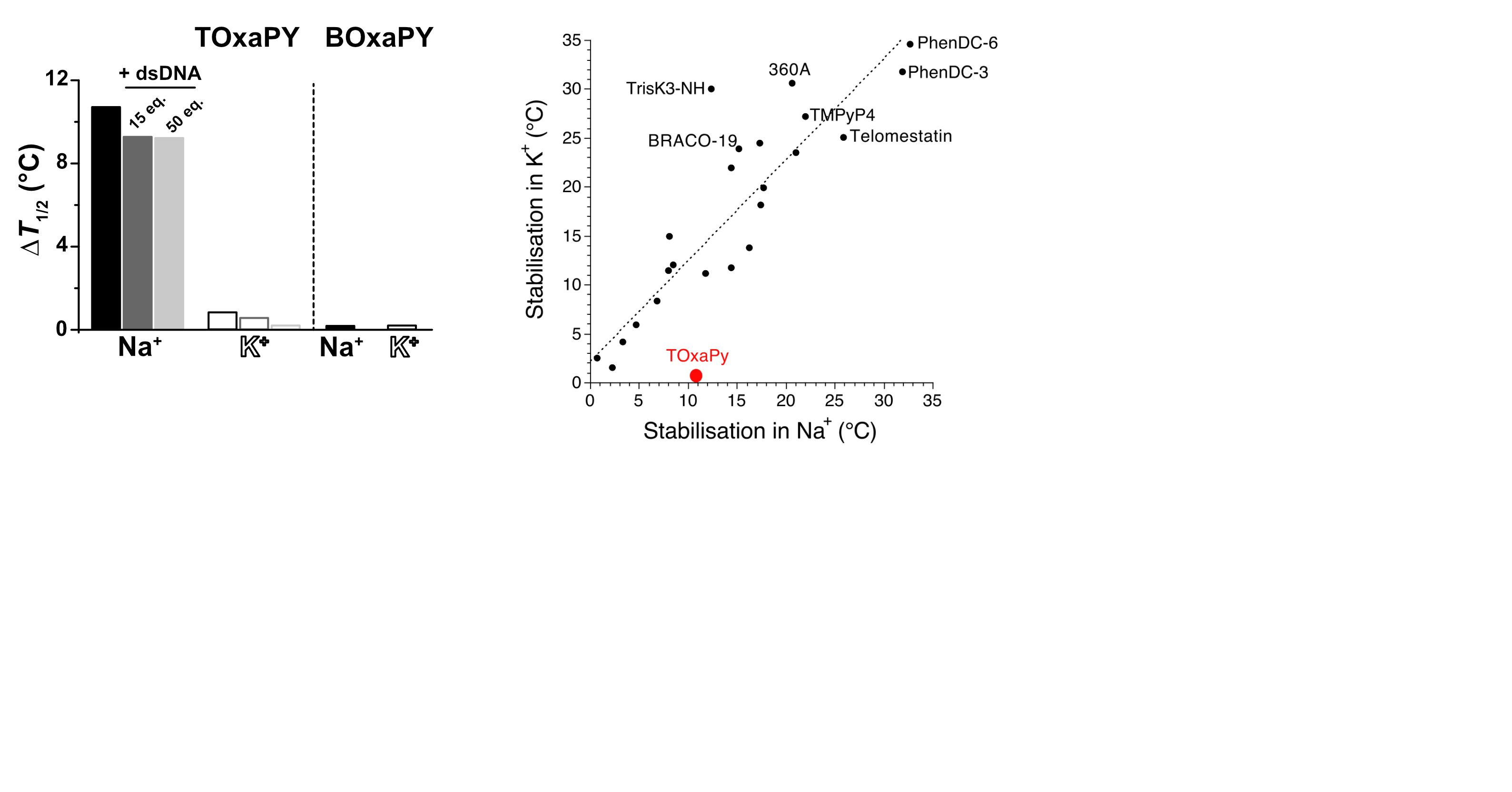
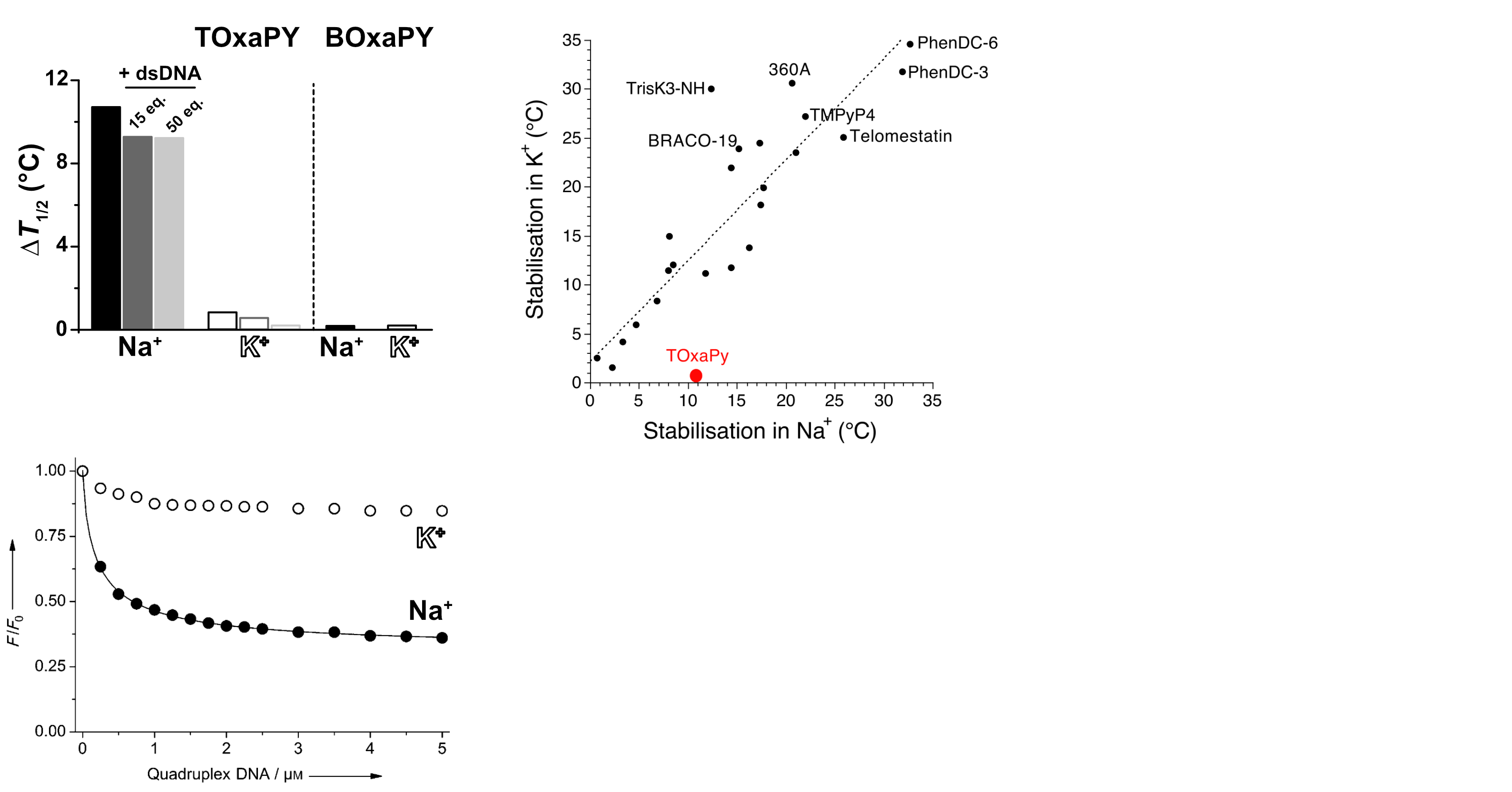
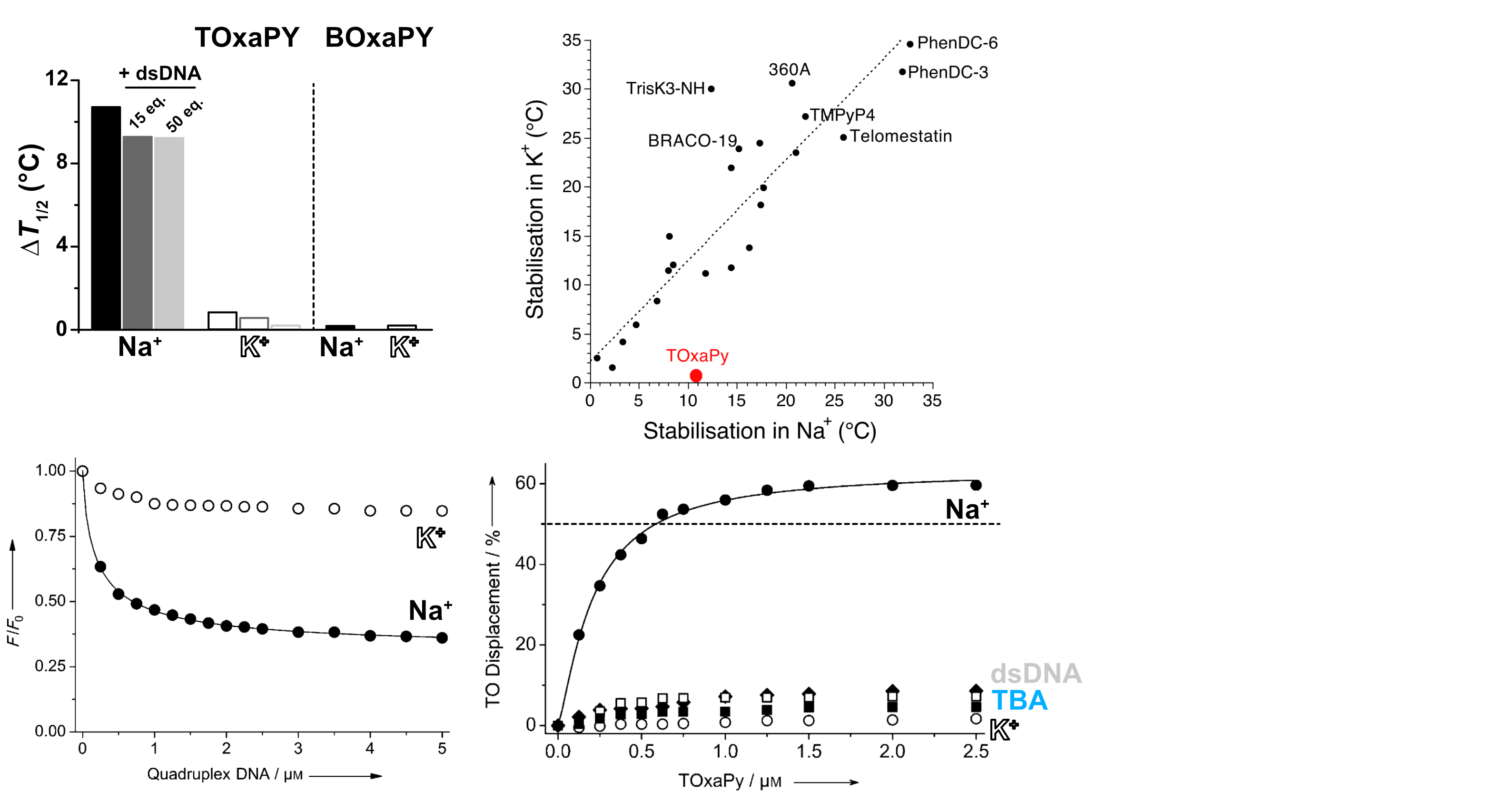
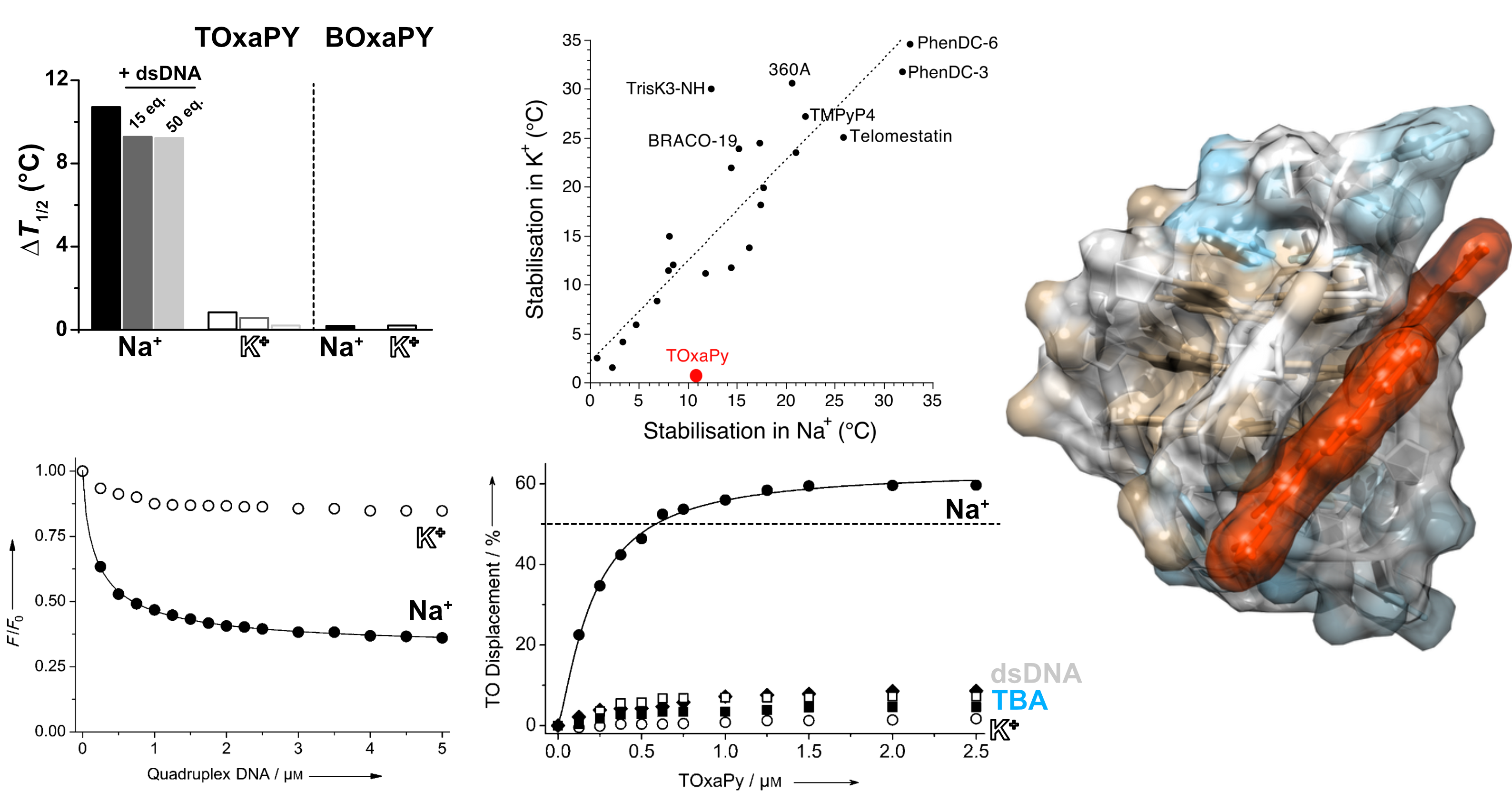
Largy, E., Guédin, A., Kabbara, A., Mergny, J.-L. et Amrane, S. Nucleic Acids Res., 2025, 53
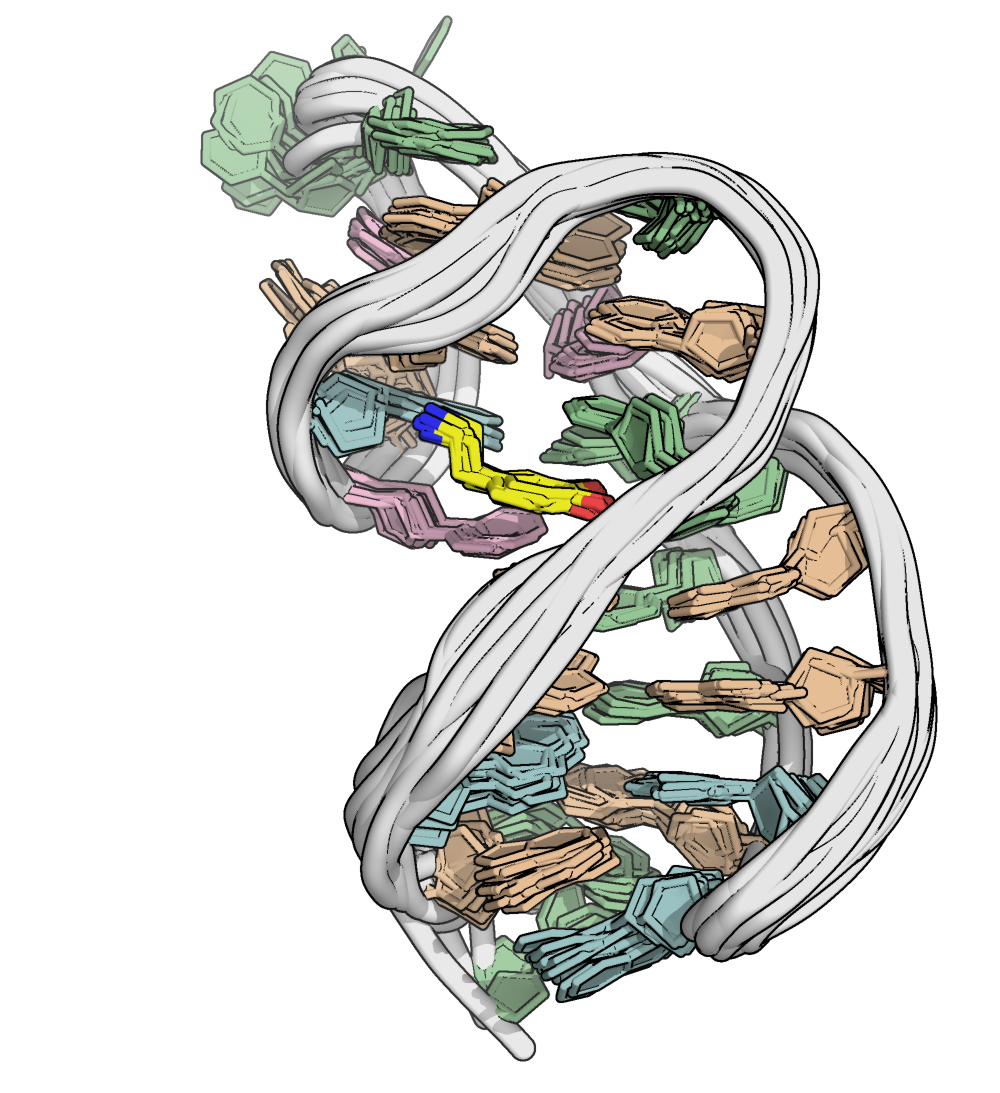
Dopamine aptamer
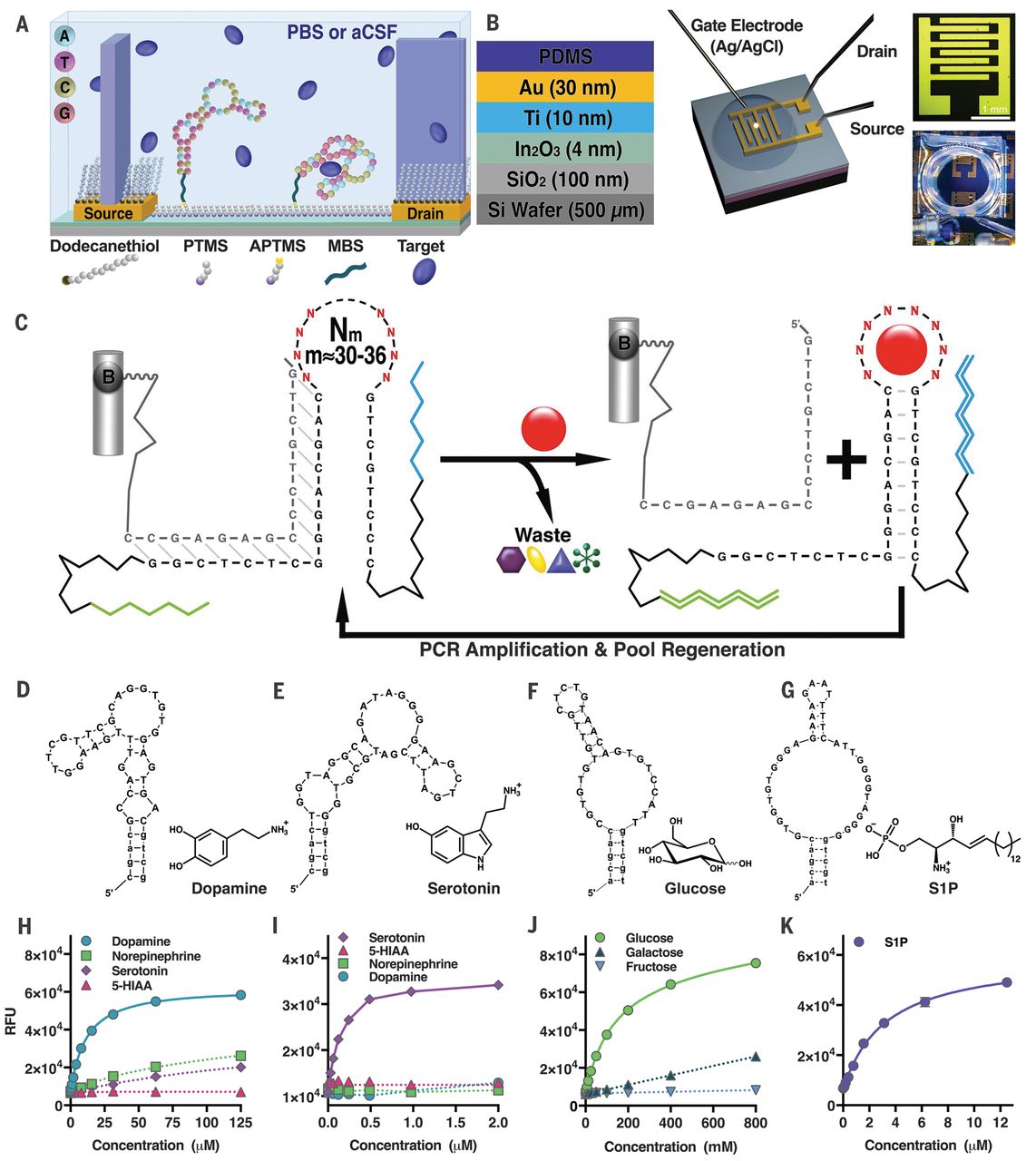
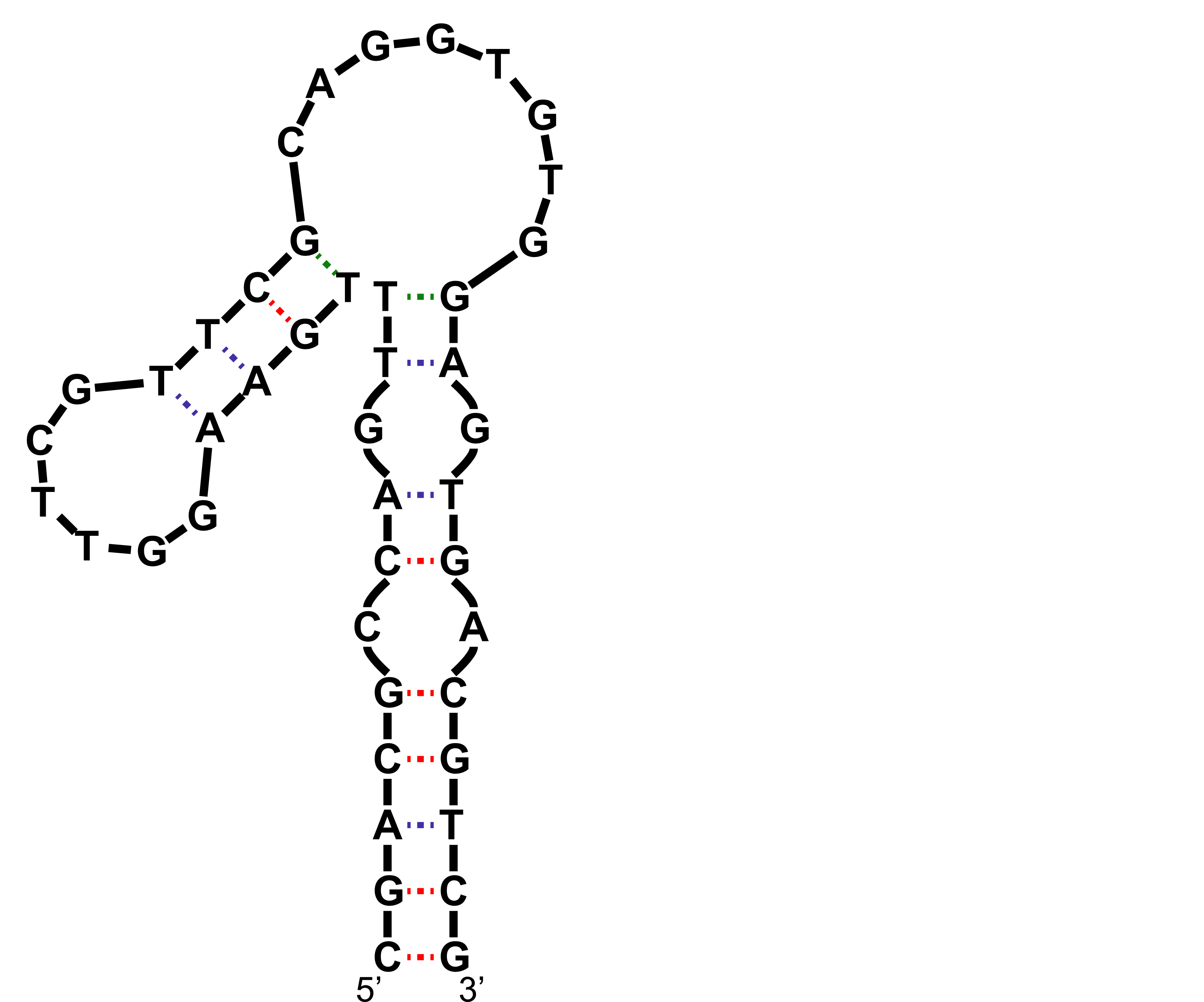
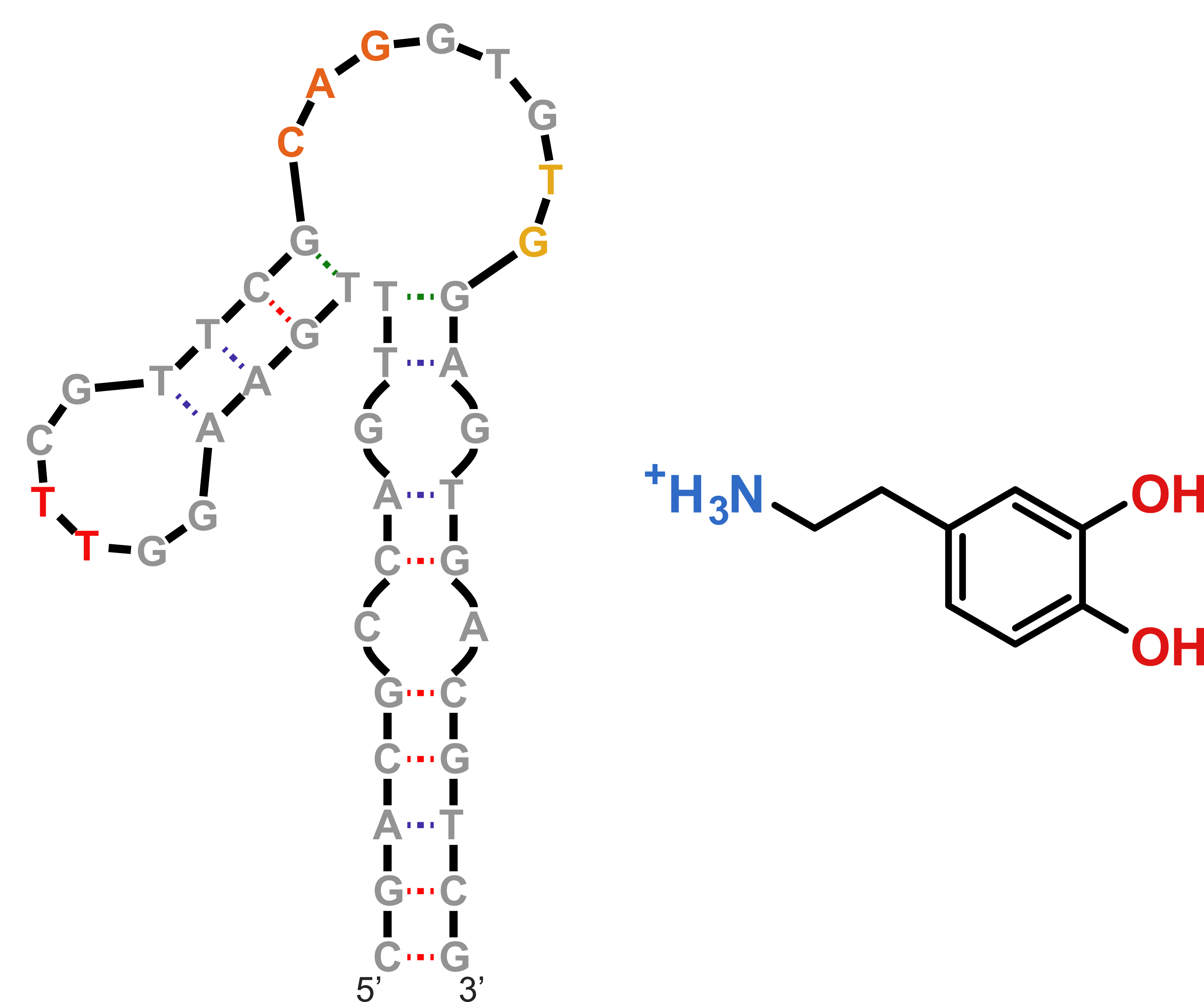
Nakatsuka N., et al., Science, 2018, 362, 319-324
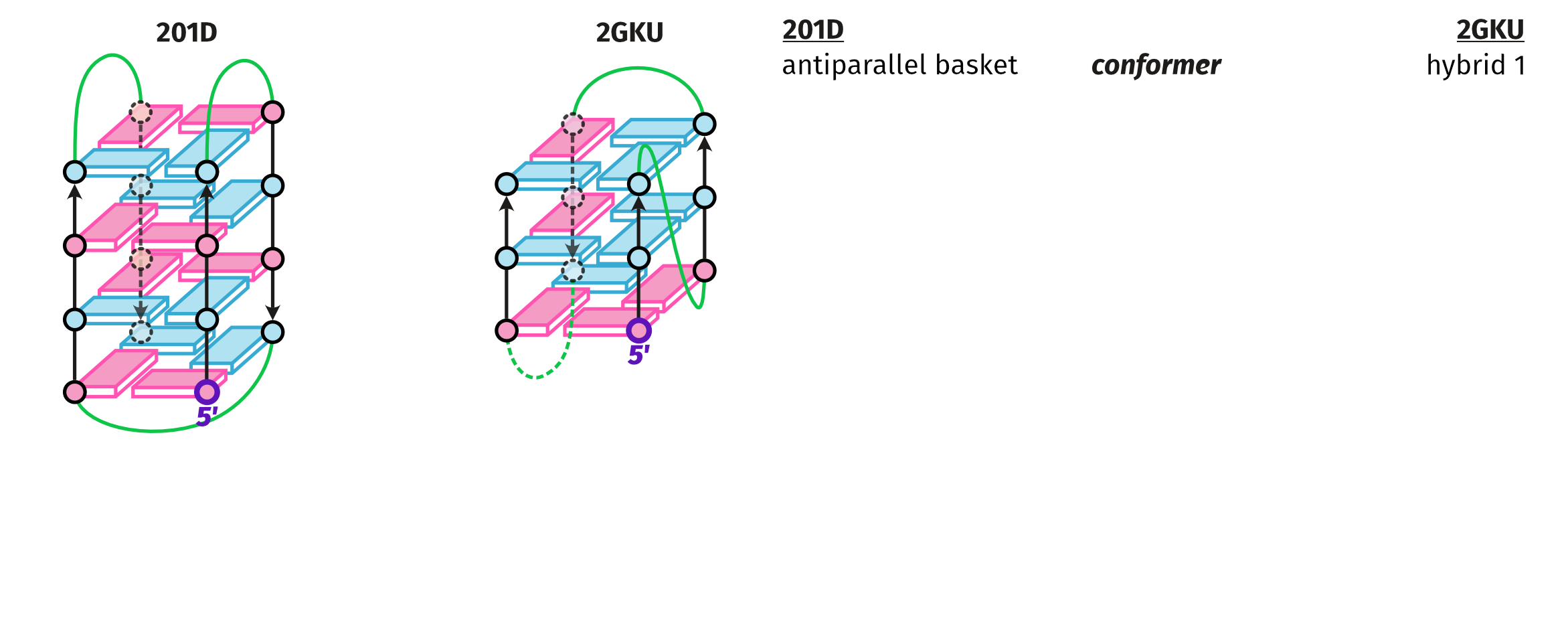
NMR provides (non-canonical !) starting models
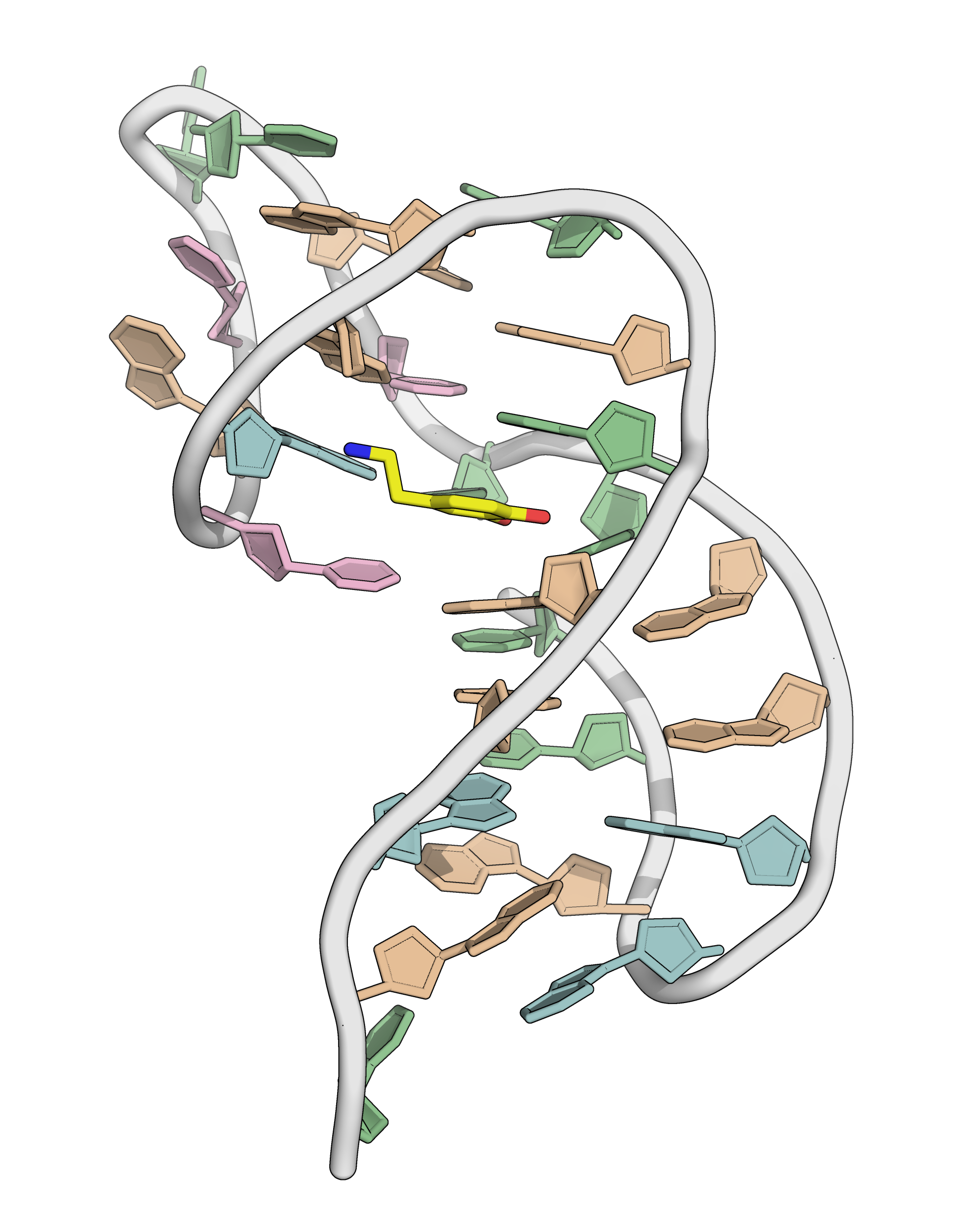

Hoi Pui Chao, E., Largy, E., Kaiyum, Y. A., Nguyen, M.-D., Vialet, B., Dauphin-Ducharme, P., Johnson, P. E., Mackereth, C.D., submitted to Nat. Chem. Biol.. BioRxiv preprint: 2025.11.27.690961
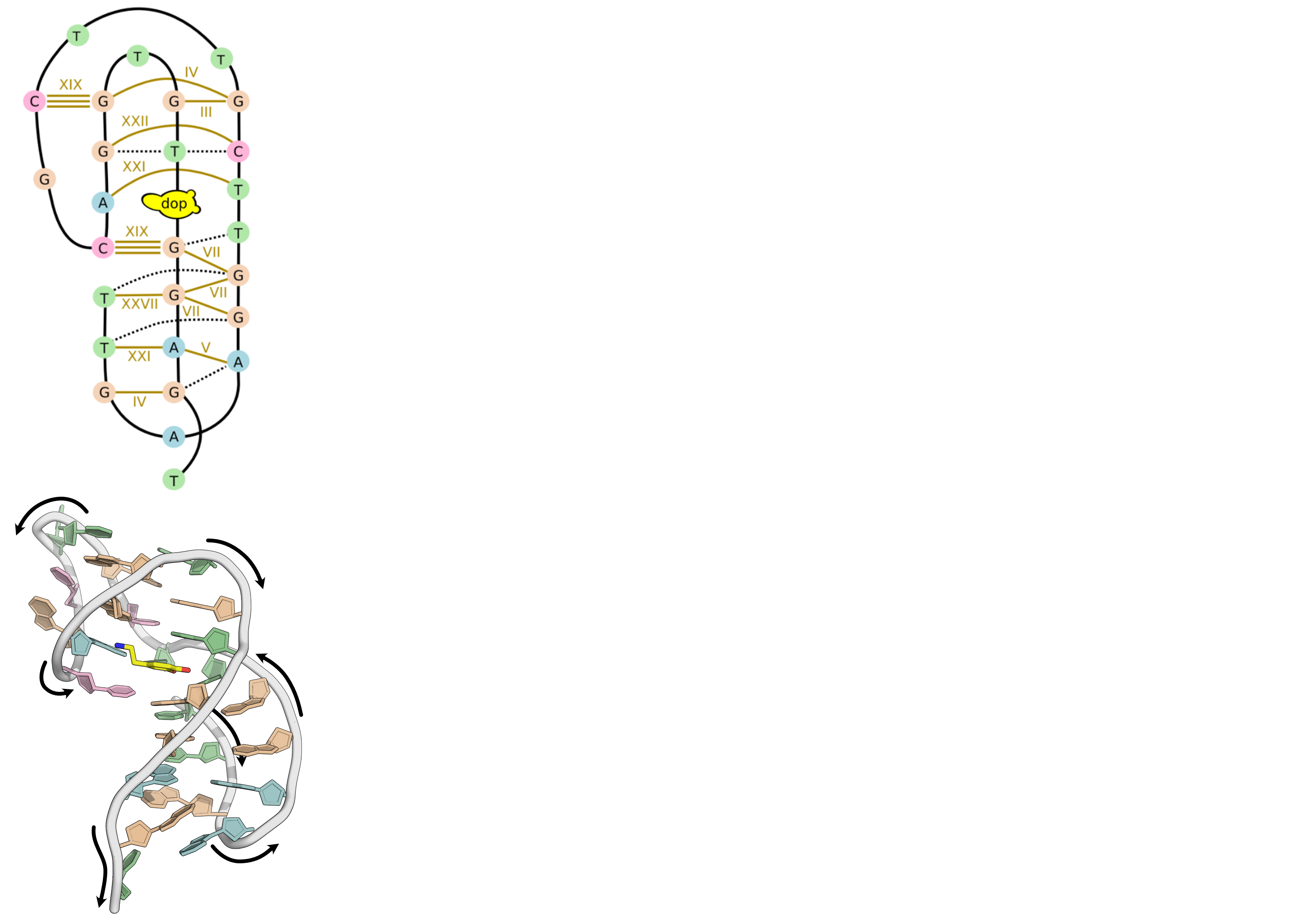
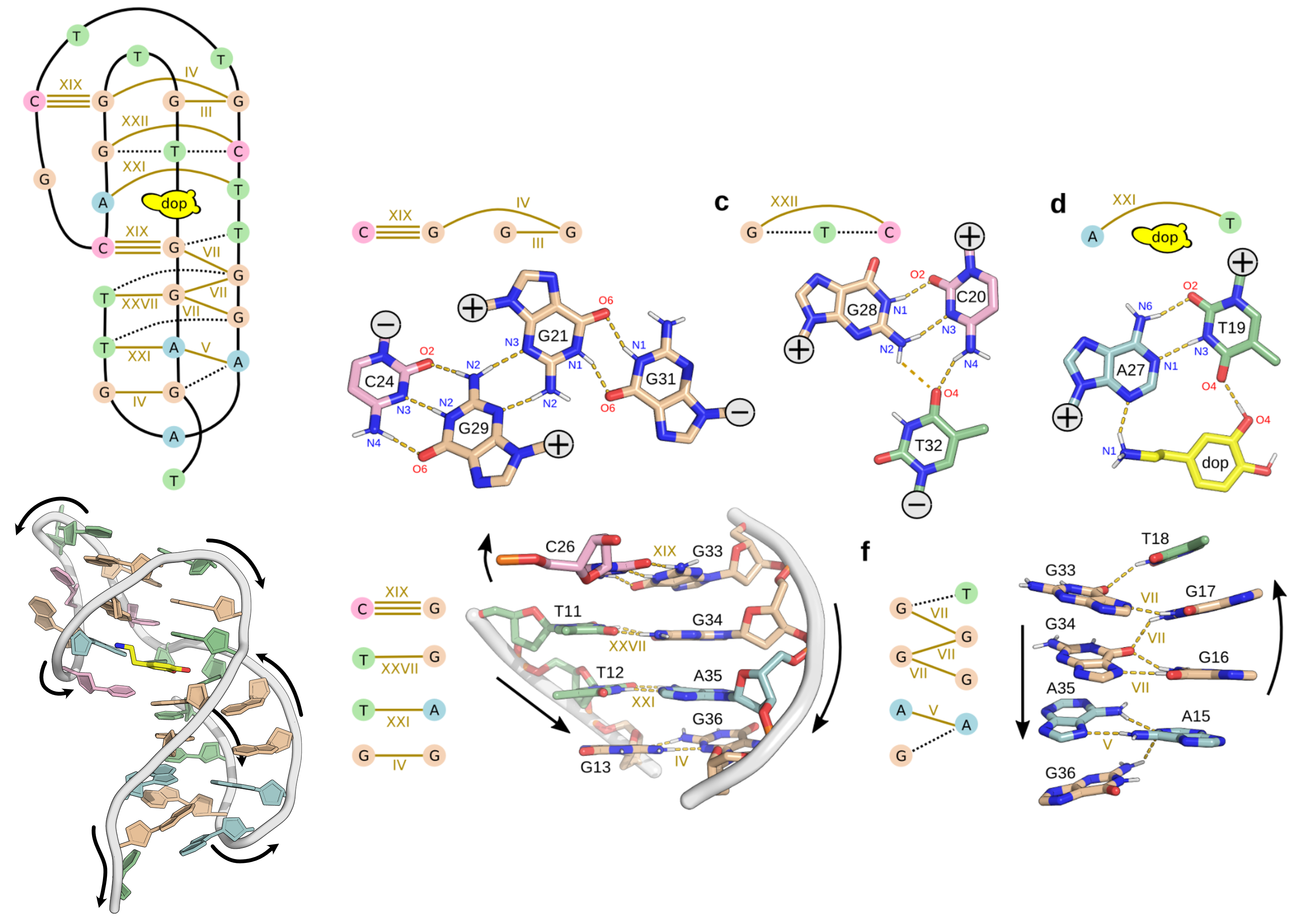
NMR provides many constraints
- DNA (806)
- intra-residue
- inter-residue
- Dopamine (67)
- intramolecular
- intermolecular
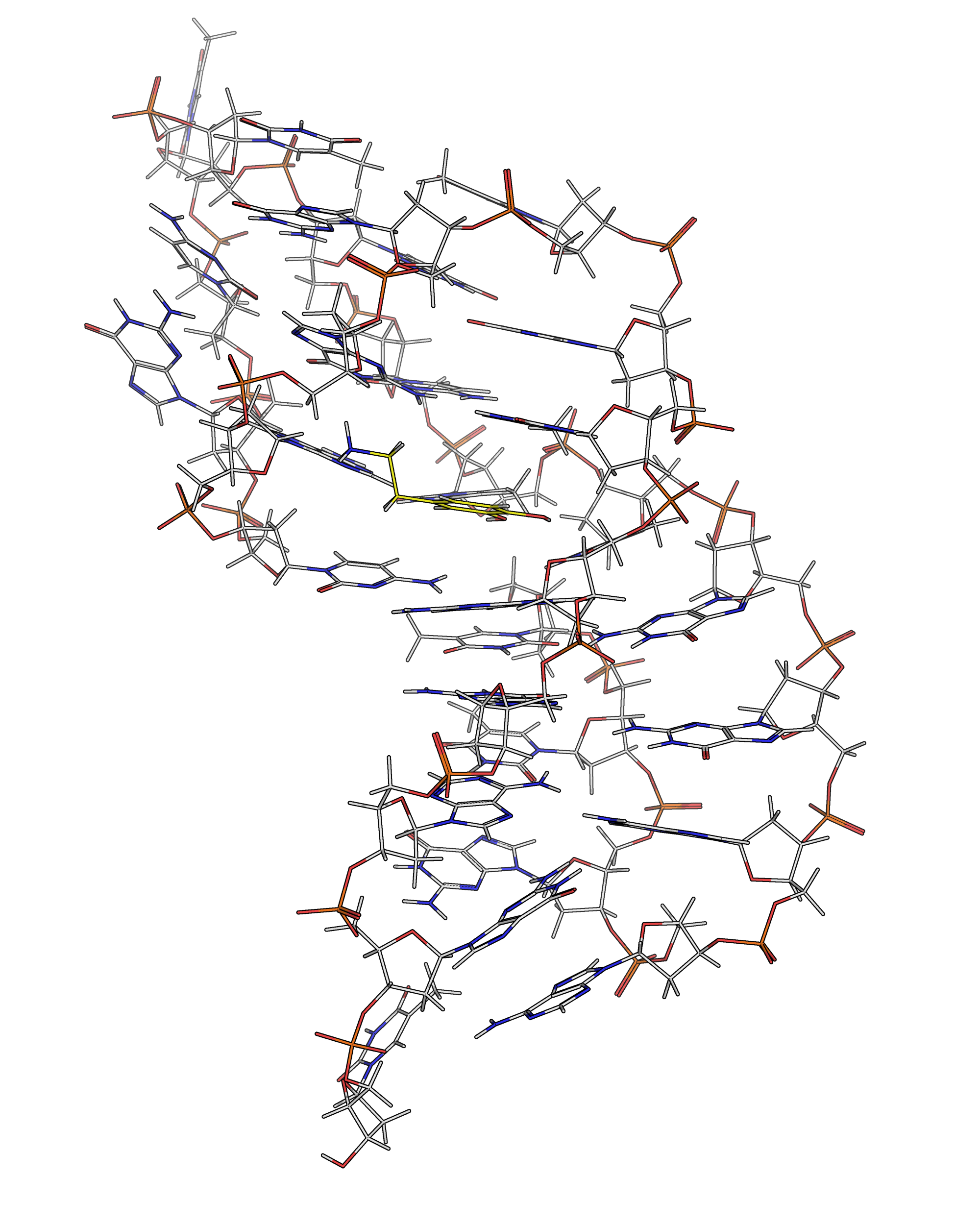
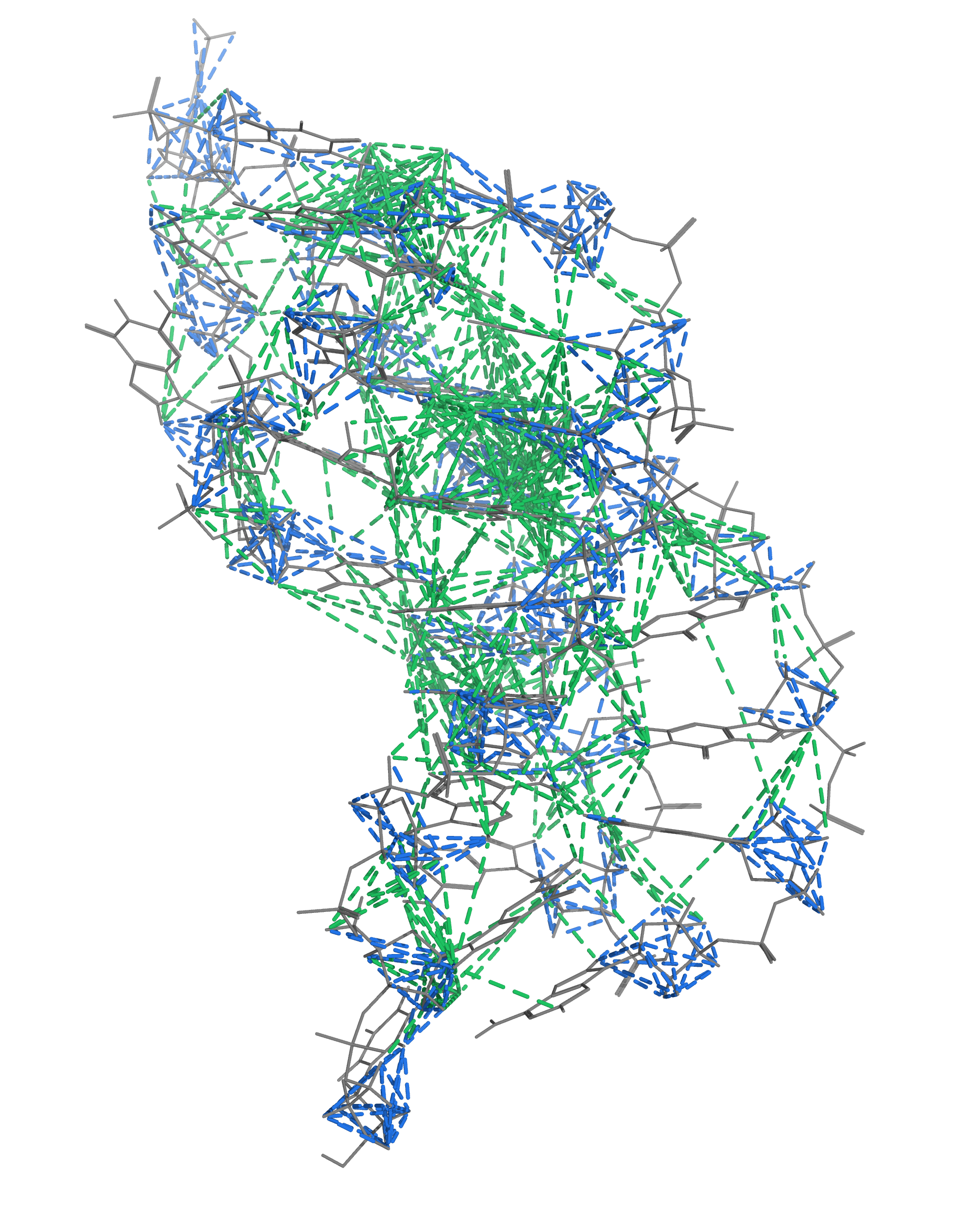
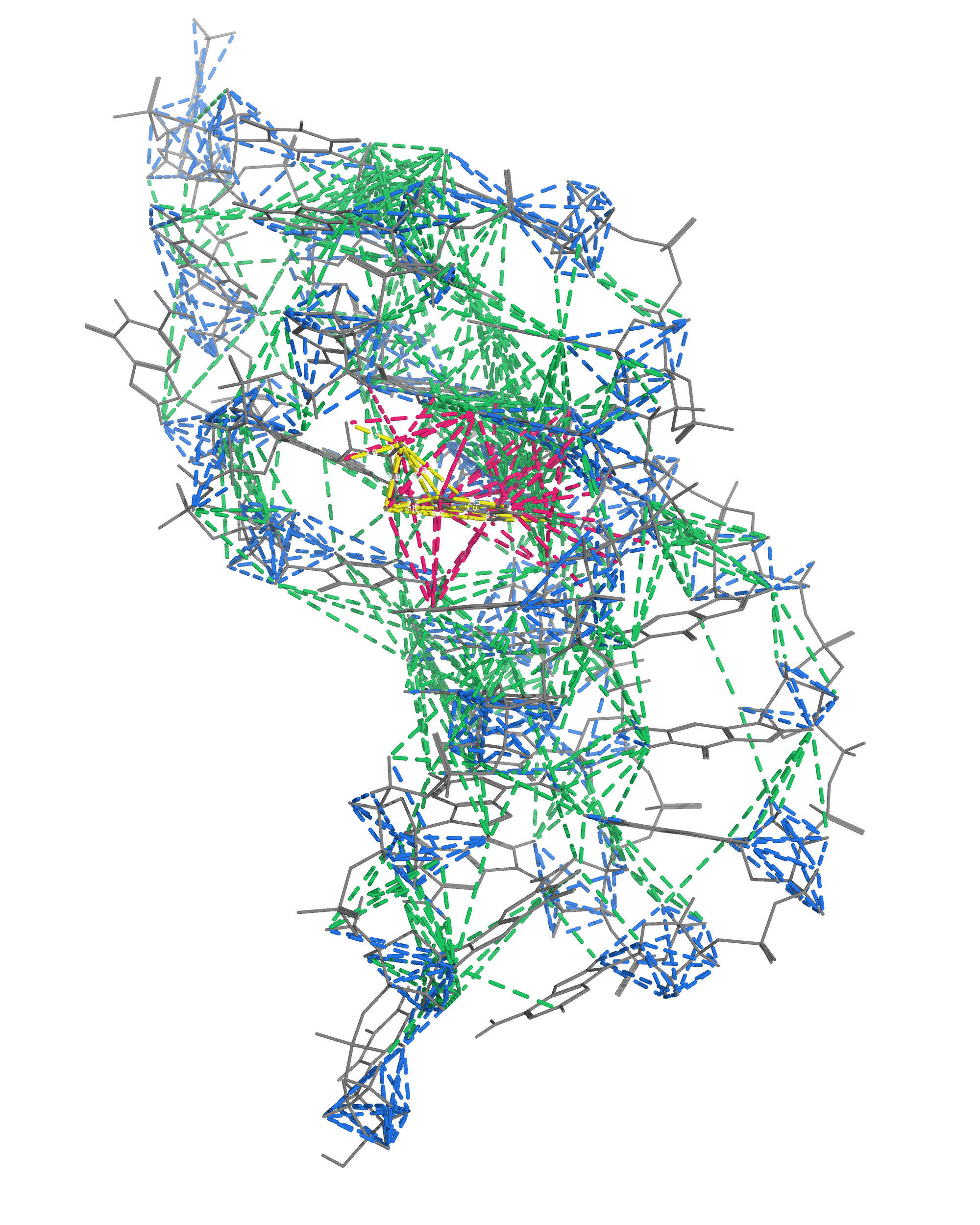

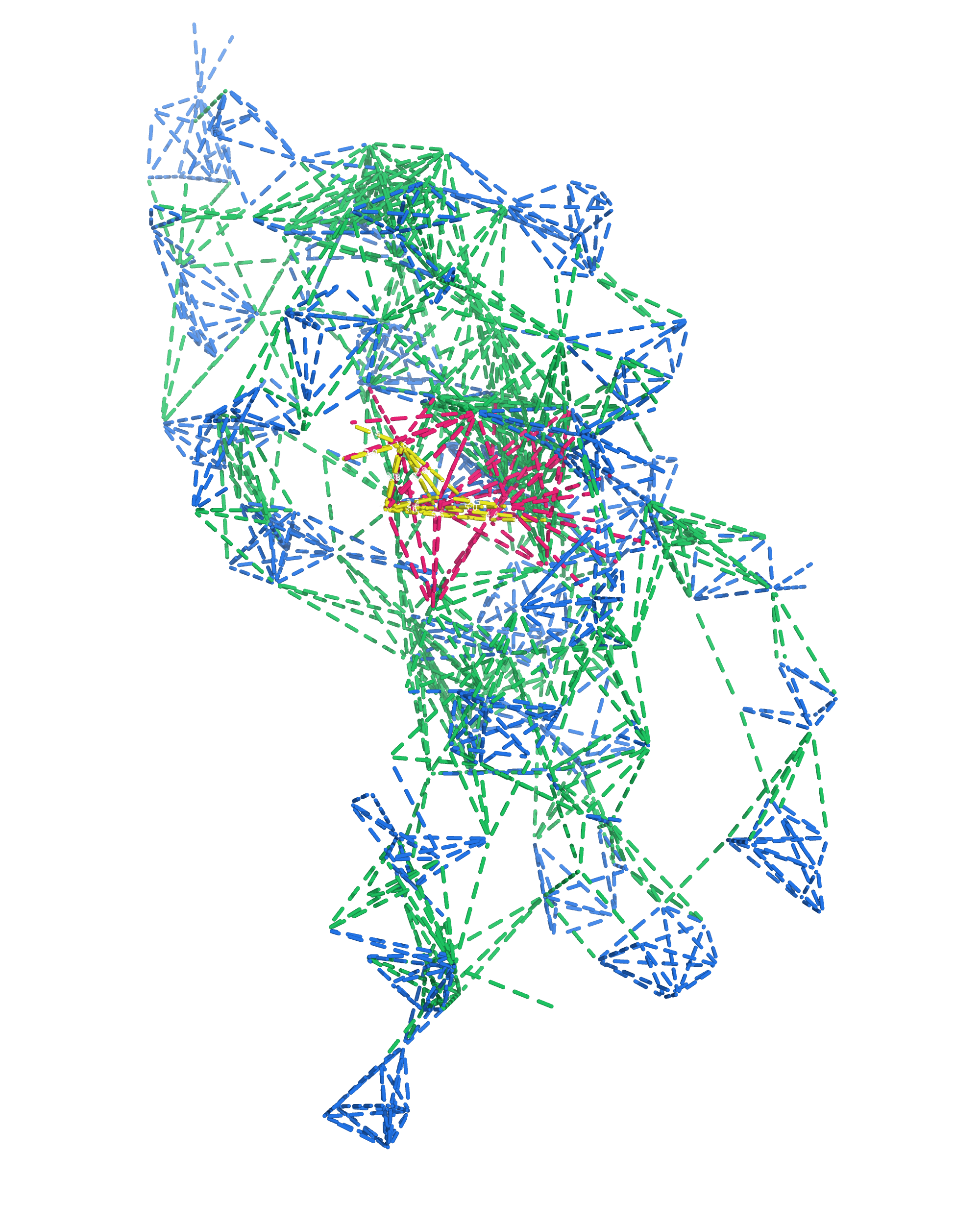

Hoi Pui Chao, E., Largy, E., Kaiyum, Y. A., Nguyen, M.-D., Vialet, B., Dauphin-Ducharme, P., Johnson, P. E., Mackereth, C.D., submitted to Nat. Chem. Biol.. BioRxiv preprint: 2025.11.27.690961
GPU-accelerated MD simulations
Hoi Pui Chao, E., Largy, E., Kaiyum, Y. A., Nguyen, M.-D., Vialet, B., Dauphin-Ducharme, P., Johnson, P. E., Mackereth, C.D., submitted to Nat. Chem. Biol.. BioRxiv preprint: 2025.11.27.690961
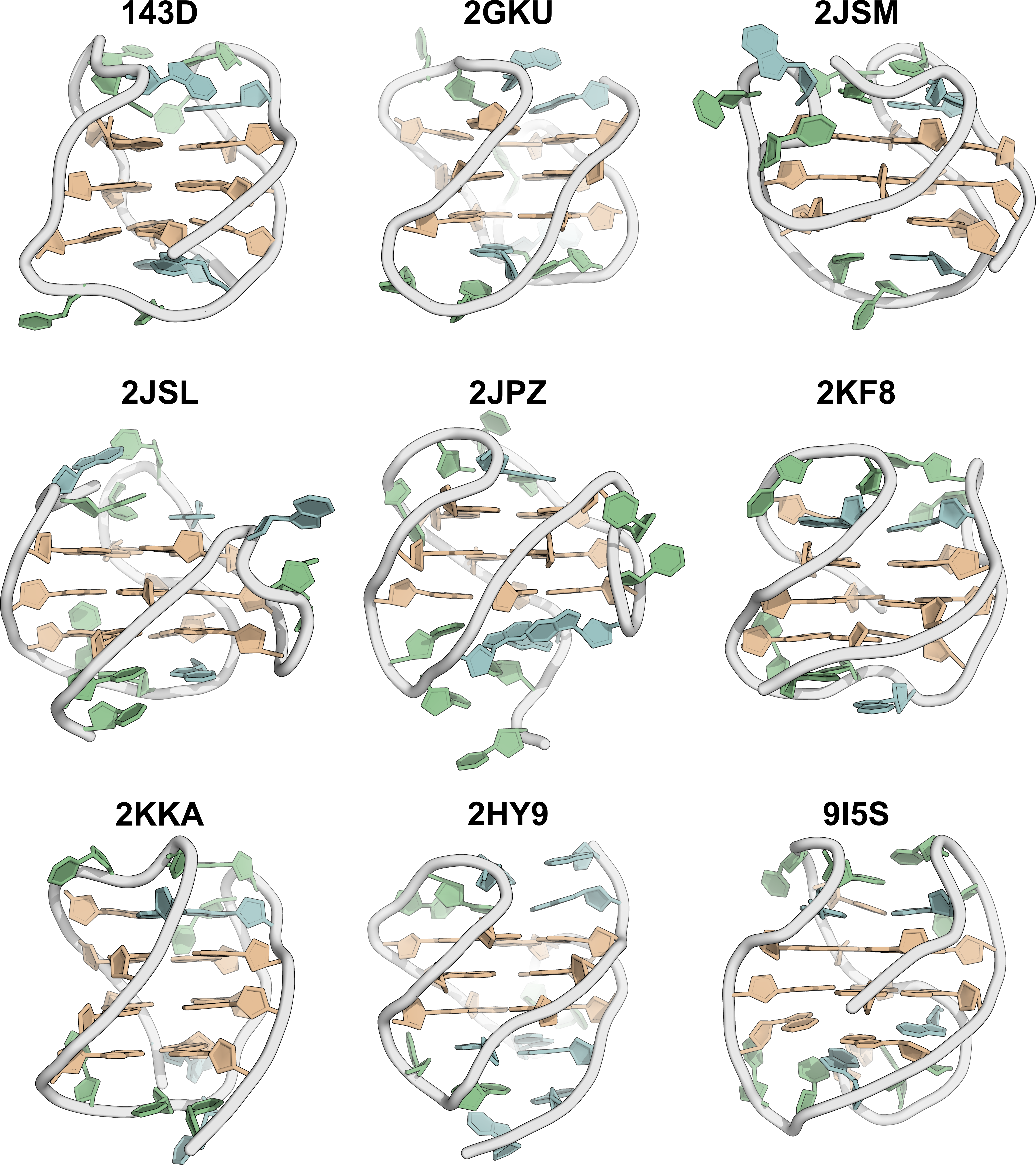
A minimized ensemble that cannot be predicted (yet)
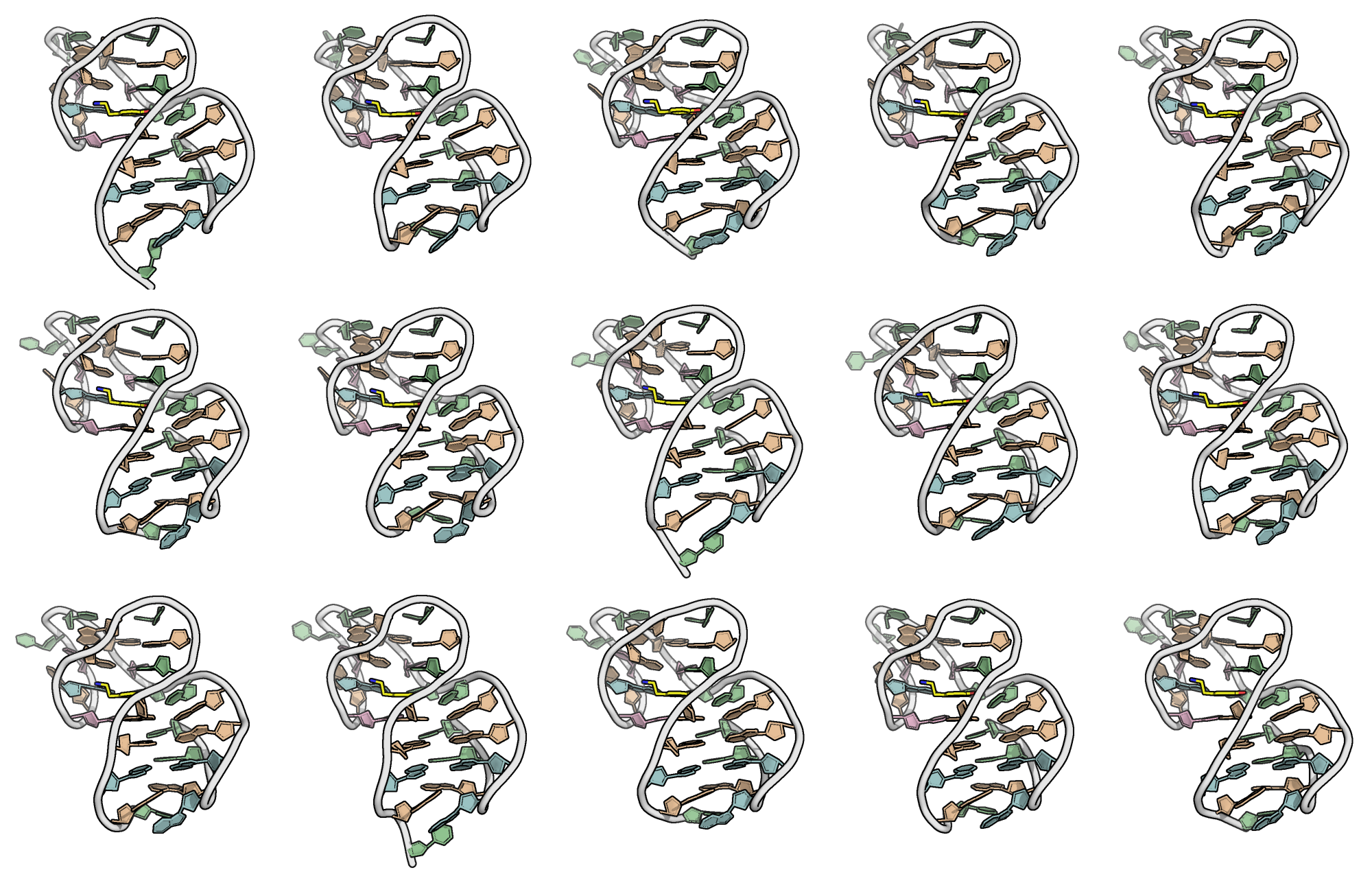

- CASP16
Critical Assessment of Techniques for Protein Structure Prediction- 107 models submitted
- 0 with RMSD < 10 Å

Kretsch R.C., et al., Proteins, 2025, prot.70043
Hoi Pui Chao, E., Largy, E., Kaiyum, Y. A., Nguyen, M.-D., Vialet, B., Dauphin-Ducharme, P., Johnson, P. E., Mackereth, C.D., submitted to Nat. Chem. Biol.. BioRxiv preprint: 2025.11.27.690961
Conclusions
Recent contributions
- Analytical method development (MS, spectroscopy), supported by:
- Open-source software for complex data analysis and database management,
- In-depth non-covalent bio-interaction characterization (G4 ligands, aptamers, HPLC), partly supported by:
- Automated and high-throughput QM/MM workflow implementation
- Nucleic Acids Res. 2025, gkaf1365
- J. Chromatogr. A 2025;1762:466395
- Nucleic Acids Res. 2025;53:gkaf953
- Proteins 2025;prot.70043
- J. Am. Chem. Soc. 2023;145:26843–26857
- Anal. Chem. 2023;acs.analchem.3c01321
- J. Am. Chem. Soc. 2023;145:498–506
- Pharmaceuticals (Basel) 2023;17:30
- Anal. Bioanal. Chem. 2022;414:6709–6721
- Pathogens 2022;11:1339
- Pharmaceuticals 2021;14:121.
- Chem. Rev. 2021;122:7720–7839
- Archiv der Pharmazie 2021;354:2000450
- Nucleic Acids Res. 2021;49:2333–2345
- Methods Mol. Biol. 2021;2271:179–188
- Methods Mol. Biol. 2021;2271:237–247
- Anal. Chem. 2020;92:4402–4410
- Biochimie 2020;179:77–84
- Nucleic Acids Res. 2019;47:2160–2168
- Nat. Commun. 2018;9:4483
Conclusions
- Selective targeting of nucleic acids by small molecules or macromolecules is still challenging
- Conformer-selective G4 ligands remain elusive (are they even needed?)
- Sequence-selectivity is not trivial
- Characterization of nucleic acid structures, complexes and dynamics:
- Requires orthogonal methods
- Can be aided by increasingly potent computational approaches that must be rooted in experimental data
- Nucleic acids therapeutics are booming but their characterization remains underdeveloped
- Limits rational design and optimization
- Methods developed at ARNA can help filling this gap
- Savoir pour prévoir, afin de pouvoir
Perspectives
- Further method development for HDX/MS of nucleic acids
- Publish HDX/IMS-MS and HDX/MS of DNA-ligand complexes
- Top-down and bottom-up approaches for increased structural resolution
- Application to protein-nucleic acid complexes
- Develop orthogonal BLI-based methods for nucleic acid complexes characterization
- Software development for custom data analysis
- Expand spectroscopic characterization of nucleic acids
- to other secondary structures, at higher throughput
- to alternative multidimensional analysis and machine learning
- Explore nucleic acid and protein folding and binding via integrated NMR/MD approaches
- Nucleic acids/small molecules
- Nucleic acids/proteins
- Small molecules/proteins
Sigl, J., Morozov, V., Wang, L., Sachs, J., Merlet, E., Largy, E., Kwon, S., Sanchez, F., Candela, L., Huet, S., Ferrand, Y., Douat, C., Mackereth, C.D., Huc, I., under review at Nat. Chem..
Conclusions
- Enriching journey through academia and industry
- Common thread: understanding biomacromolecule folding and interactions
- From synthetic chemistry to biophysics and computational approaches
- From fundamental research to applied science
Moi, si je devais résumer ma vie aujourd’hui avec vous, je dirais que c’est d’abord des rencontres […] Et c’est assez curieux de se dire que les hasards, les rencontres forgent une destinée. Parce que quand on a le goût de la chose, quand on a le goût de la chose bien faite, le beau geste, parfois on ne trouve pas l’interlocuteur en face je dirais, le miroir qui vous aide à avancer.
– Otis
Thank you
- Marie-Paule Teulade-Fichou @ UMR 176/Institut Curie
- Florian Hamon, Daniela Verga & Anton Granzhan
- David Perrin @ UBC
- Abid Hasan & Wenbo Liu
- Jean-Louis Mergny @ U869/IECB
- Aurore Guédin-Beaurepaire, Carmelo Di Primo
- Arnaud Delobel @ Quality Assistance
- Gery Van Vyncht, Fabrice Cantais & Anicet Catrain
- Alain Beck (Pierre Fabre)
- Chris Hosfield & Sergei Saveliev (Promega)
- David Lascoux, Guillaume Béchade & Matthew Laubner (Waters)
- Ose Immunotherapeutics
- Valérie Gabelica @ U1212/IECB
- Frédéric Rosu
- Adrien Marchand & Valentina D’Atri
- Steven Daly, Nina Khristenko, Anirban Ghosh, Desbamita Ghosh & Dominika Strzelecka
- Matthieu Ranz & Alexander König
- Cédric Mesmin & Jérôme Haustant (Merck)
- Cameron Mackereth @ U1212/BBS
- Samir Amrane, Pierre Bonnafous, Gilmar Salgado
- Karen Gaudin @ UFR Pharmacie
- Bruno Alies, Ludivine Ferey, Alexandra Gaubert, Marie-Hélène Langlois & Thu-Van Nguyen
- Funding
- Novartis, Bourse M2 Fédération Gay-Lussac
- Institut Curie/CNRS, Bourse de docteur ingénieur
- NSERC/CRSNG, Banting postdoctoral fellowship
- Union Européenne : ERDF funding
- INSERM
- University of Bordeaux, fédération TecSan
- ANR, JCJC
- All the students
- Platforms
- Emily Seo, UBC
- Corinne Buré & Frédéric Rosu, IECB
- Estelle Morvan & Axelle Grelard, IECB
- Xavier Montagutelli, Pierre Gay, U. Limoges
- Personnels administratifs
- Laetitia Cointement, Sandra Lavenant, Malika Vloeberghs
- Tous les responsables de…
- L’unité U1212
- Ma famille, des deux côtés de l’Atlantique
Toutes celles et ceux que j’ai oubliés
Thanks to all collaborators
Find yourself @ network.largy.fr